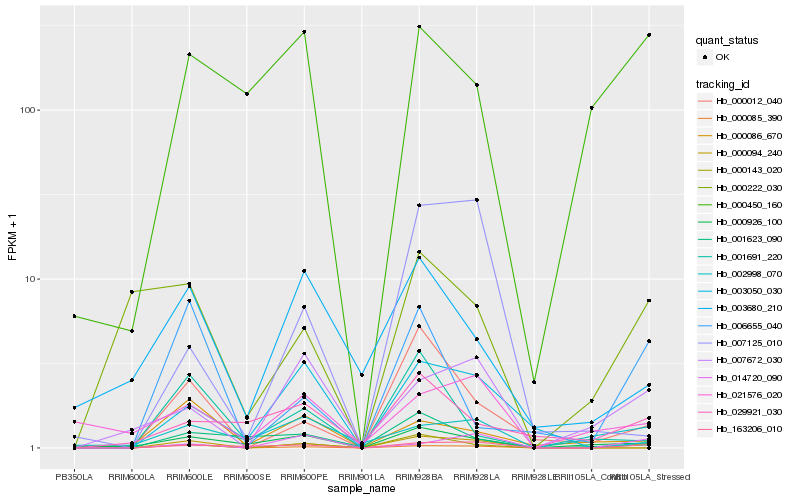
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 2 |
Hb_163206_010 |
0.2272813146 |
- |
- |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 3 |
Hb_000012_040 |
0.2535445583 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 4 |
Hb_000926_100 |
0.2929774844 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 5 |
Hb_000085_390 |
0.2992764653 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000143_020 |
0.3130267867 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_007672_030 |
0.3239984479 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 8 |
Hb_003050_030 |
0.3252713618 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000222_030 |
0.3267155111 |
- |
- |
- |
| 10 |
Hb_006655_040 |
0.3355168067 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 11 |
Hb_001623_090 |
0.3433818085 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 12 |
Hb_014720_090 |
0.3470635745 |
- |
- |
- |
| 13 |
Hb_003680_210 |
0.3475003869 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 14 |
Hb_001691_220 |
0.3475996607 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 15 |
Hb_000086_670 |
0.3485712234 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 16 |
Hb_021576_020 |
0.3506151254 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 17 |
Hb_000450_160 |
0.3517964882 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 18 |
Hb_029921_030 |
0.3557647209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002998_070 |
0.3574548469 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 20 |
Hb_007125_010 |
0.3595684293 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |