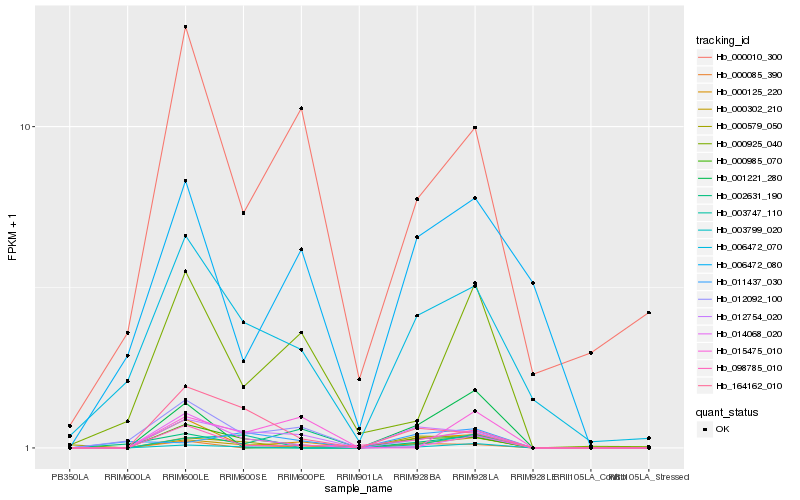
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000985_070 |
0.1972819104 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 3 |
Hb_012754_020 |
0.2101931931 |
- |
- |
PREDICTED: uncharacterized protein LOC104229837 [Nicotiana sylvestris] |
| 4 |
Hb_098785_010 |
0.2198212597 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 5 |
Hb_000125_220 |
0.2230965989 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 13 [Jatropha curcas] |
| 6 |
Hb_003799_020 |
0.224494025 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 7 |
Hb_000925_040 |
0.278819346 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_006472_070 |
0.2794537139 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 9 |
Hb_000085_390 |
0.301901774 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_003747_110 |
0.3048075515 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 11 |
Hb_000579_050 |
0.3186784121 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 12 |
Hb_001221_280 |
0.3202725749 |
- |
- |
- |
| 13 |
Hb_012092_100 |
0.3226604397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000010_300 |
0.3247604903 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_014068_020 |
0.3263391293 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 16 |
Hb_164162_010 |
0.3270384237 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 17 |
Hb_006472_080 |
0.3275715822 |
- |
- |
- |
| 18 |
Hb_002631_190 |
0.3411502083 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 19 |
Hb_015475_010 |
0.3430084378 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 20 |
Hb_011437_030 |
0.3442318515 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |