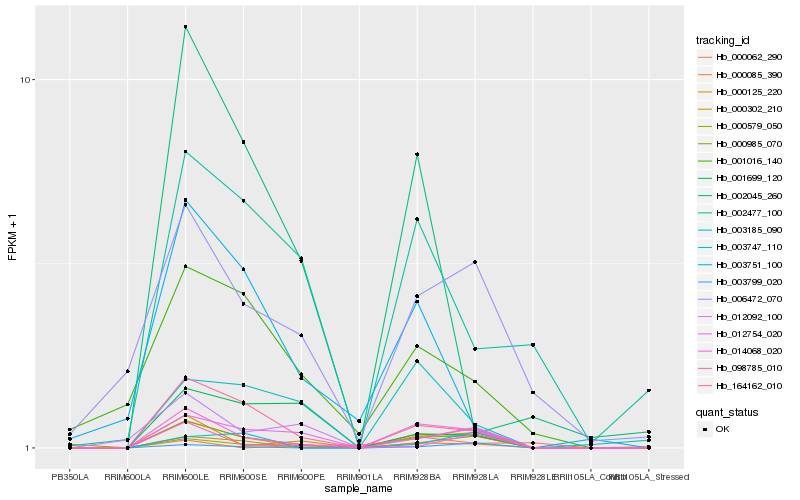
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000985_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 2 |
Hb_164162_010 |
0.1570343892 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 3 |
Hb_012754_020 |
0.1617101354 |
- |
- |
PREDICTED: uncharacterized protein LOC104229837 [Nicotiana sylvestris] |
| 4 |
Hb_000302_210 |
0.1972819104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000125_220 |
0.203518257 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 13 [Jatropha curcas] |
| 6 |
Hb_014068_020 |
0.2361375252 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 7 |
Hb_098785_010 |
0.2529155759 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_006472_070 |
0.2649371336 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 9 |
Hb_012092_100 |
0.2765468448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001016_140 |
0.2874774449 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 11 |
Hb_000062_290 |
0.2926642056 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |
| 12 |
Hb_003799_020 |
0.294255983 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 13 |
Hb_003747_110 |
0.3075893895 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 14 |
Hb_003185_090 |
0.3083713712 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 15 |
Hb_002477_100 |
0.3124503953 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 16 |
Hb_002045_260 |
0.3128645348 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 17 |
Hb_000579_050 |
0.3129143773 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 18 |
Hb_001699_120 |
0.3151706383 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 19 |
Hb_003751_100 |
0.3153267785 |
- |
- |
Amino acid permease 2 [Glycine soja] |
| 20 |
Hb_000085_390 |
0.3193744438 |
- |
- |
unnamed protein product [Vitis vinifera] |