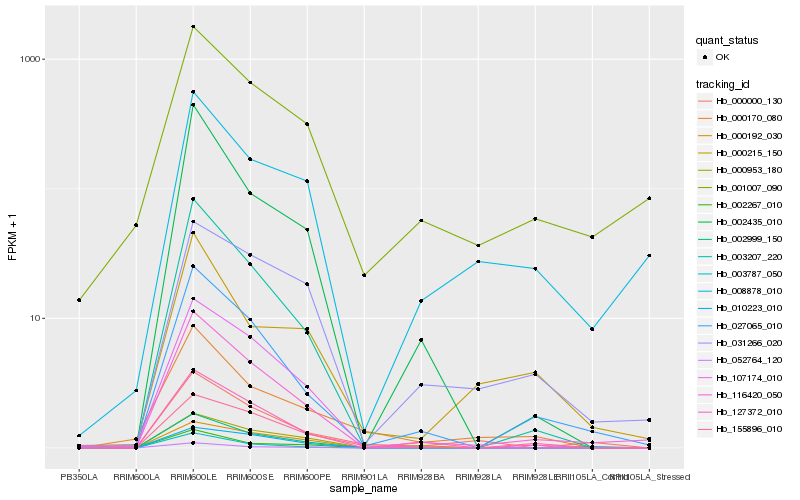
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_130 |
0.0 |
- |
- |
Disease resistance-responsive family protein [Theobroma cacao] |
| 2 |
Hb_116420_050 |
0.1763434214 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 3 |
Hb_027065_010 |
0.1896652373 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_000170_080 |
0.1905745124 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 5 |
Hb_002999_150 |
0.1976649767 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 6 |
Hb_003787_050 |
0.198090281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_107174_010 |
0.2019654704 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003207_220 |
0.2035765596 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 9 |
Hb_127372_010 |
0.2078930454 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 10 |
Hb_001007_090 |
0.2085203835 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 11 |
Hb_002435_010 |
0.2132059257 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 12 |
Hb_000953_180 |
0.2166232743 |
- |
- |
- |
| 13 |
Hb_052764_120 |
0.2169772688 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 14 |
Hb_008878_010 |
0.2172843727 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 15 |
Hb_000192_030 |
0.217806705 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 16 |
Hb_155896_010 |
0.2184397185 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 17 |
Hb_031266_020 |
0.2190117082 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010223_010 |
0.2203834844 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_000215_150 |
0.2218913048 |
- |
- |
PREDICTED: uncharacterized protein LOC100249186 [Vitis vinifera] |
| 20 |
Hb_002267_010 |
0.2227988688 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |