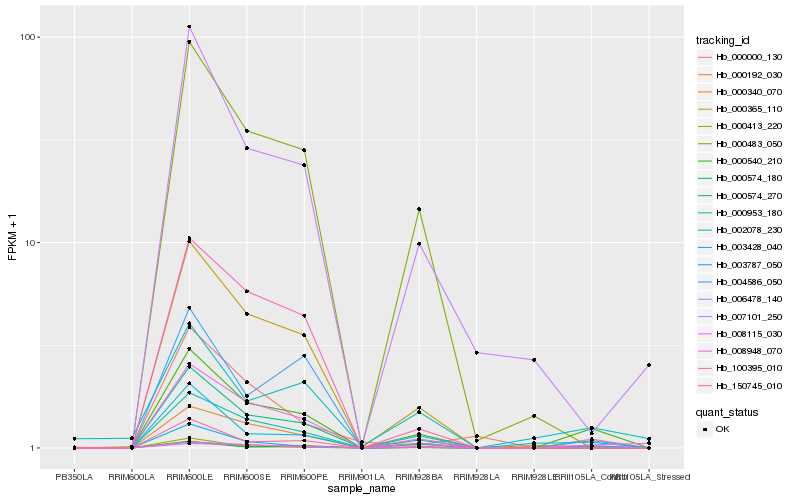
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008115_030 |
0.1676675499 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 3 |
Hb_003787_050 |
0.1712643462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000365_110 |
0.1867511921 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 5 |
Hb_003428_040 |
0.1870759633 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000574_270 |
0.188190831 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 7 |
Hb_000483_050 |
0.2006921048 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 8 |
Hb_000953_180 |
0.2007493192 |
- |
- |
- |
| 9 |
Hb_150745_010 |
0.2137499838 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 10 |
Hb_007101_250 |
0.2170267248 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 11 |
Hb_004586_050 |
0.2224824334 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 12 |
Hb_000192_030 |
0.2241064761 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 13 |
Hb_008948_070 |
0.2247121596 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_100395_010 |
0.2265217683 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000340_070 |
0.2278204359 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 16 |
Hb_000413_220 |
0.227877471 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 17 |
Hb_006478_140 |
0.2305710407 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 18 |
Hb_000000_130 |
0.231201619 |
- |
- |
Disease resistance-responsive family protein [Theobroma cacao] |
| 19 |
Hb_000574_180 |
0.2313834016 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 20 |
Hb_002078_230 |
0.2338080878 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |