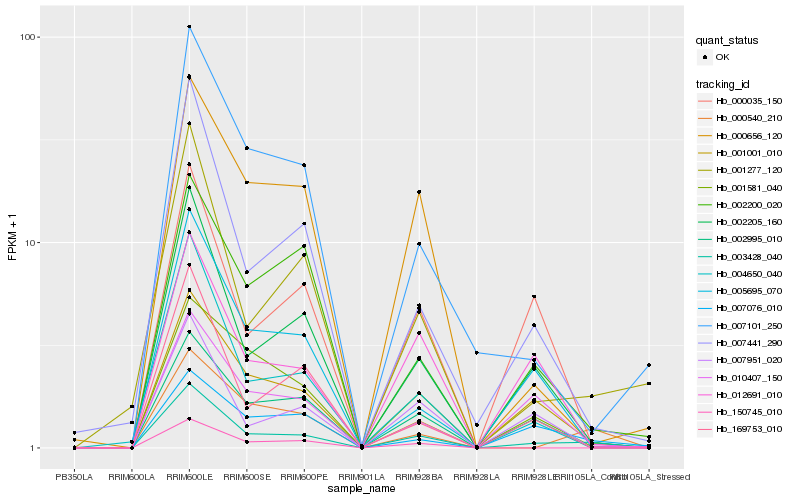
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003428_040 |
0.0 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002205_160 |
0.1535040908 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 3 |
Hb_001001_010 |
0.1685898367 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_004650_040 |
0.1721992335 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 5 |
Hb_007441_290 |
0.1723218429 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 6 |
Hb_001277_120 |
0.1849107887 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 7 |
Hb_000540_210 |
0.1870759633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002995_010 |
0.1905093722 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 9 |
Hb_010407_150 |
0.1938018222 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 10 |
Hb_005695_070 |
0.1959309828 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 11 |
Hb_007951_020 |
0.1963213569 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 12 |
Hb_012691_010 |
0.1979273192 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 13 |
Hb_150745_010 |
0.2011372685 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 14 |
Hb_000035_150 |
0.2015698961 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 15 |
Hb_007101_250 |
0.2032184483 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 16 |
Hb_169753_010 |
0.2035672407 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 17 |
Hb_000656_120 |
0.2043609362 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 18 |
Hb_001581_040 |
0.2045435675 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_002200_020 |
0.2051523392 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 20 |
Hb_007076_010 |
0.2052090378 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |