| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
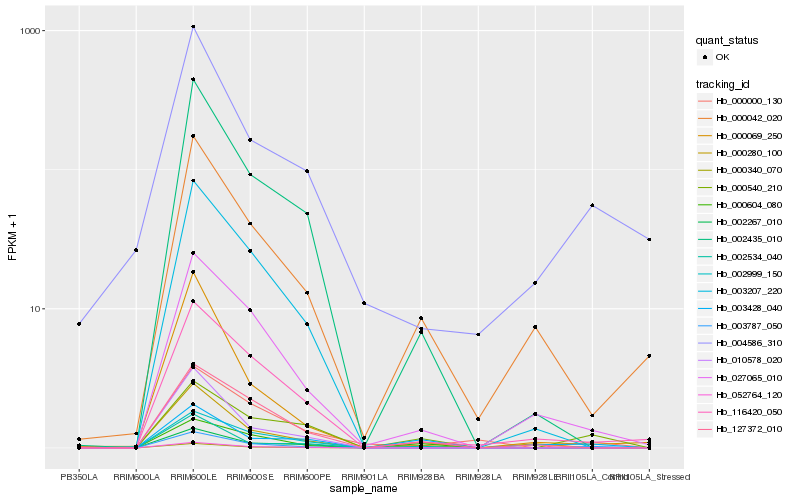
Hb_003787_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000540_210 |
0.1712643462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_027065_010 |
0.1791605113 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 4 |
Hb_002435_010 |
0.1928082188 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 5 |
Hb_116420_050 |
0.1980680291 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 6 |
Hb_000000_130 |
0.198090281 |
- |
- |
Disease resistance-responsive family protein [Theobroma cacao] |
| 7 |
Hb_000340_070 |
0.1994672715 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 8 |
Hb_004586_310 |
0.2096773452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003207_220 |
0.2099173685 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 10 |
Hb_052764_120 |
0.2100728562 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 11 |
Hb_127372_010 |
0.2139595358 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 12 |
Hb_000280_100 |
0.2150805652 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 13 |
Hb_010578_020 |
0.2154308021 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000069_250 |
0.2162191889 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 15 |
Hb_000042_020 |
0.216650774 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002534_040 |
0.2215620456 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 17 |
Hb_002999_150 |
0.2233592874 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 18 |
Hb_000604_080 |
0.2241395091 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 19 |
Hb_003428_040 |
0.2265035935 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002267_010 |
0.2272941989 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |