| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
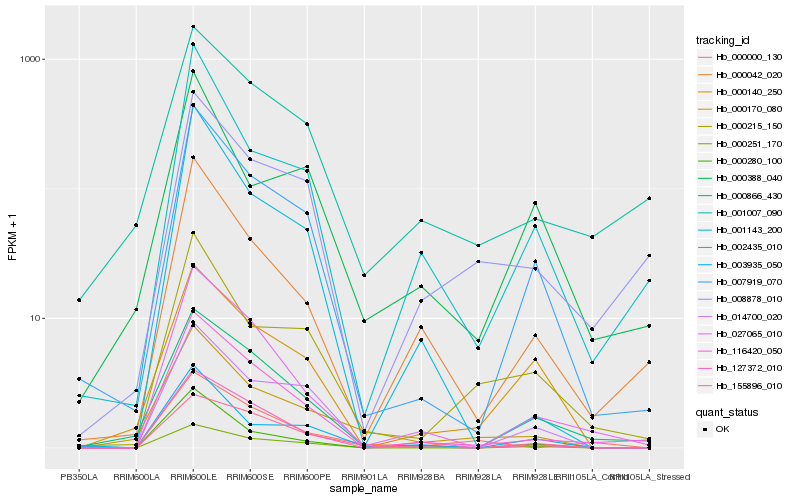
Hb_000170_080 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 2 |
Hb_000215_150 |
0.169289 |
- |
- |
PREDICTED: uncharacterized protein LOC100249186 [Vitis vinifera] |
| 3 |
Hb_007919_070 |
0.1789175765 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 4 |
Hb_001007_090 |
0.1845081316 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15 [Jatropha curcas] |
| 5 |
Hb_003935_050 |
0.1846338848 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000140_250 |
0.1903704676 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 7 |
Hb_000000_130 |
0.1905745124 |
- |
- |
Disease resistance-responsive family protein [Theobroma cacao] |
| 8 |
Hb_000388_040 |
0.1925733779 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 9 |
Hb_155896_010 |
0.1937071423 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 10 |
Hb_001143_200 |
0.1949306862 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 11 |
Hb_000280_100 |
0.1980035082 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 12 |
Hb_116420_050 |
0.1987096417 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 13 |
Hb_002435_010 |
0.19949935 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 14 |
Hb_000042_020 |
0.2003887643 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000251_170 |
0.2010827614 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000866_430 |
0.2045352181 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 17 |
Hb_014700_020 |
0.2051637417 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 18 |
Hb_008878_010 |
0.2061040228 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Populus euphratica] |
| 19 |
Hb_027065_010 |
0.2067952269 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 20 |
Hb_127372_010 |
0.207357462 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |