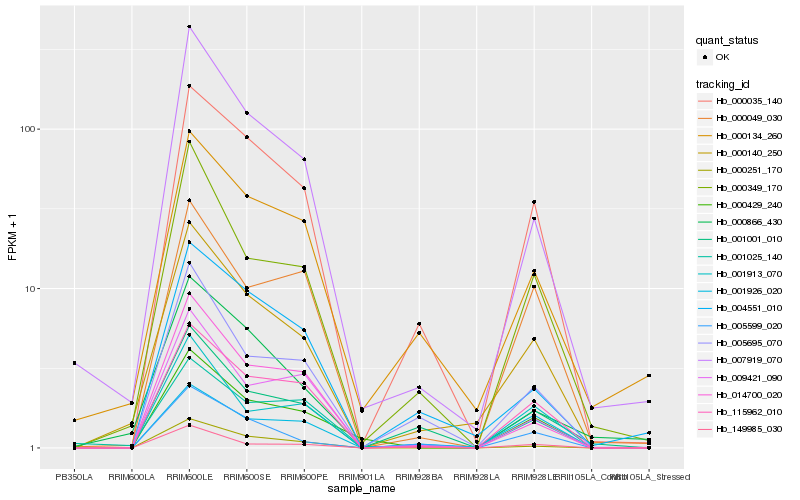
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_250 |
0.0 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 2 |
Hb_000035_140 |
0.1260895283 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_000349_170 |
0.1331185288 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_115962_010 |
0.1399870246 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 5 |
Hb_007919_070 |
0.1412981919 |
- |
- |
hypothetical protein POPTR_0005s28110g [Populus trichocarpa] |
| 6 |
Hb_005695_070 |
0.1470538913 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 7 |
Hb_000134_260 |
0.1501331025 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_005599_020 |
0.1522757925 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 9 |
Hb_149985_030 |
0.1576282204 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 10 |
Hb_001913_070 |
0.1594852508 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 11 |
Hb_004551_010 |
0.1613301541 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 12 |
Hb_000251_170 |
0.16311643 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001001_010 |
0.163878198 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 14 |
Hb_000866_430 |
0.1656243103 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 15 |
Hb_014700_020 |
0.1661438862 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 16 |
Hb_000429_240 |
0.1685768398 |
- |
- |
PREDICTED: uncharacterized protein LOC105645185 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001025_140 |
0.1699037595 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 18 |
Hb_009421_090 |
0.1711192356 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 19 |
Hb_000049_030 |
0.1744252931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001926_020 |
0.1766716631 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |