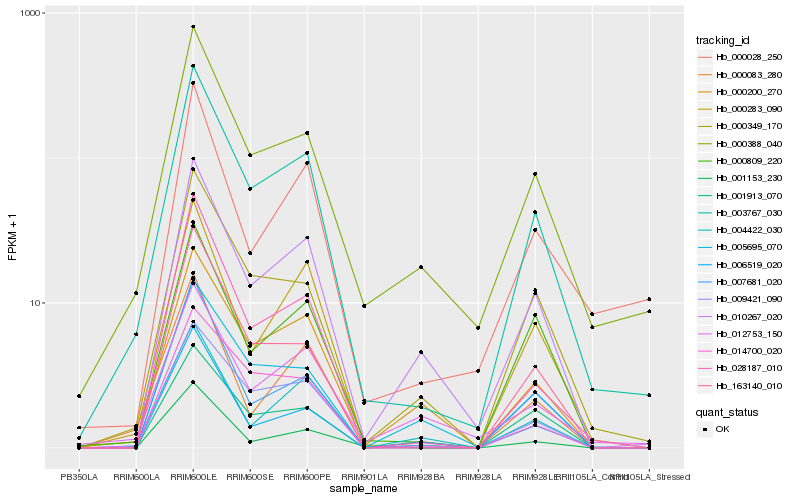
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003767_030 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 2 |
Hb_000388_040 |
0.1033434664 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 3 |
Hb_000349_170 |
0.111841233 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_009421_090 |
0.1125516717 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 5 |
Hb_000283_090 |
0.1199006424 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_010267_020 |
0.1215332361 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 7 |
Hb_000809_220 |
0.1217981747 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |
| 8 |
Hb_028187_010 |
0.1277719186 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 9 |
Hb_000200_270 |
0.1279594234 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_163140_010 |
0.1284661826 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 11 |
Hb_007681_020 |
0.1303994825 |
- |
- |
transferase, putative [Ricinus communis] |
| 12 |
Hb_001913_070 |
0.1343647186 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 13 |
Hb_000083_280 |
0.1368005988 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_012753_150 |
0.141666527 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 15 |
Hb_006519_020 |
0.142026985 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_004422_030 |
0.1437991321 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 17 |
Hb_014700_020 |
0.1439685845 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 18 |
Hb_005695_070 |
0.1440076092 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 19 |
Hb_000028_250 |
0.1449375706 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 20 |
Hb_001153_230 |
0.1455252779 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |