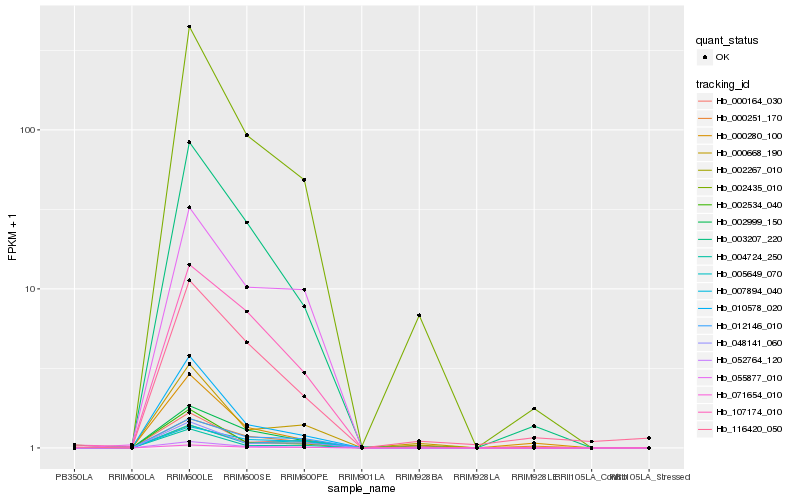
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 2 |
Hb_002267_010 |
0.0458844131 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 3 |
Hb_010578_020 |
0.0692335748 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002435_010 |
0.0757887781 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 5 |
Hb_003207_220 |
0.0774889696 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 6 |
Hb_002999_150 |
0.082260363 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 7 |
Hb_048141_060 |
0.0834895939 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 8 |
Hb_071654_010 |
0.0937477834 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 9 |
Hb_004724_250 |
0.1003360149 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000164_030 |
0.1096205088 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 11 |
Hb_012146_010 |
0.135736455 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 12 |
Hb_055877_010 |
0.1394909557 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_107174_010 |
0.1423114601 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_007894_040 |
0.1457787067 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000668_190 |
0.1471046486 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 16 |
Hb_000251_170 |
0.1564959035 |
- |
- |
leucine-rich repeat transmembrane protein kinase, putative [Ricinus communis] |
| 17 |
Hb_005649_070 |
0.1575736318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_116420_050 |
0.1580301493 |
- |
- |
PREDICTED: ACT domain-containing protein ACR1 [Jatropha curcas] |
| 19 |
Hb_002534_040 |
0.1599159791 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 20 |
Hb_000280_100 |
0.1602244043 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |