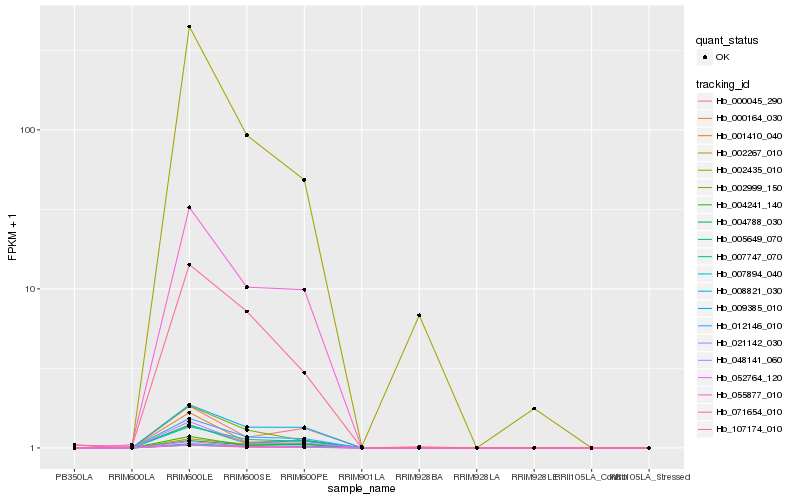
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071654_010 |
0.0 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 2 |
Hb_012146_010 |
0.0446174379 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 3 |
Hb_055877_010 |
0.0495816223 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_002267_010 |
0.0531831952 |
- |
- |
hypothetical protein SORBIDRAFT_01g040370 [Sorghum bicolor] |
| 5 |
Hb_007894_040 |
0.059191377 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007747_070 |
0.0813967461 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 7 |
Hb_002999_150 |
0.0866263631 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 8 |
Hb_048141_060 |
0.0875727185 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 9 |
Hb_052764_120 |
0.0937477834 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 10 |
Hb_005649_070 |
0.0956843192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_021142_030 |
0.1061617174 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 12 |
Hb_000164_030 |
0.1064571181 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 13 |
Hb_004241_140 |
0.1084345759 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 14 |
Hb_009385_010 |
0.1110229502 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 15 |
Hb_008821_030 |
0.1125192774 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 16 |
Hb_001410_040 |
0.1188396972 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 17 |
Hb_002435_010 |
0.1191704745 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 18 |
Hb_000045_290 |
0.1213866697 |
- |
- |
- |
| 19 |
Hb_004788_030 |
0.1259467627 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_107174_010 |
0.1264367533 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |