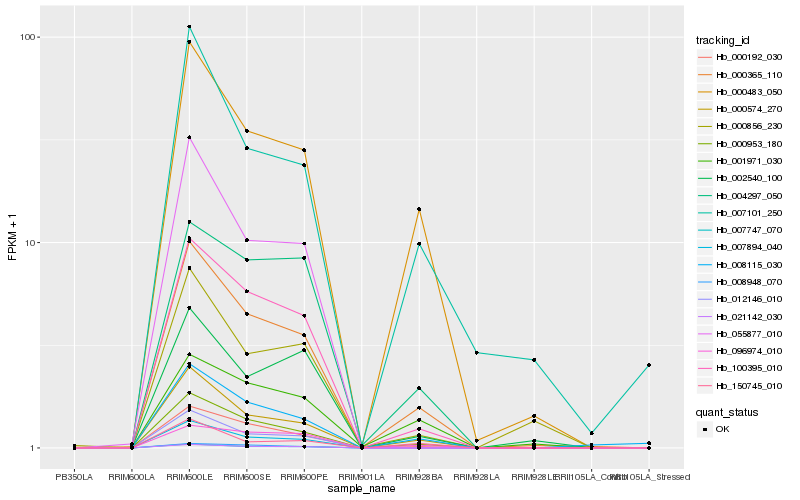
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_110 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 2 |
Hb_000953_180 |
0.0652365251 |
- |
- |
- |
| 3 |
Hb_000574_270 |
0.0659134362 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_000483_050 |
0.0885427107 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 5 |
Hb_100395_010 |
0.0944130289 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000192_030 |
0.0994537125 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 7 |
Hb_008948_070 |
0.1104941339 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_008115_030 |
0.1261293314 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 9 |
Hb_007101_250 |
0.13823222 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 10 |
Hb_001971_030 |
0.141696471 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 11 |
Hb_007894_040 |
0.1425994312 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002540_100 |
0.1428620175 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_000856_230 |
0.1443686863 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 14 |
Hb_012146_010 |
0.1451370843 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 15 |
Hb_096974_010 |
0.1452363176 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 16 |
Hb_007747_070 |
0.1464701009 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 17 |
Hb_004297_050 |
0.1480604201 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_021142_030 |
0.1486090434 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 19 |
Hb_055877_010 |
0.1510327525 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_150745_010 |
0.1523652919 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |