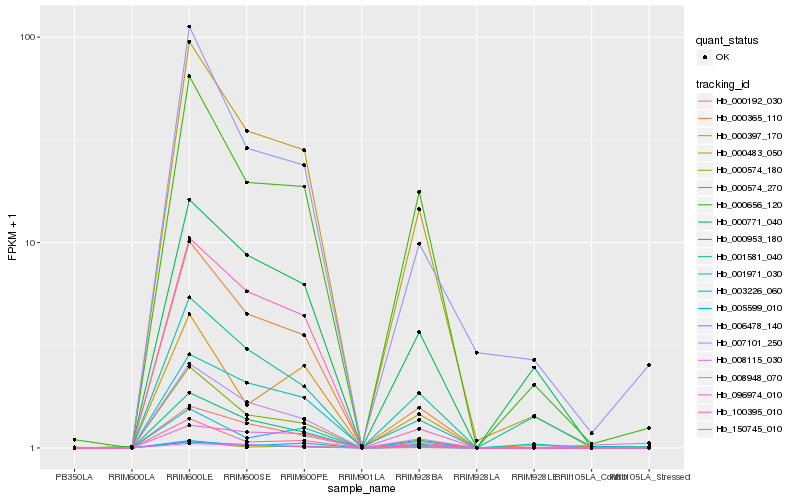
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_050 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 2 |
Hb_000365_110 |
0.0885427107 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 3 |
Hb_000574_270 |
0.0966459725 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_001971_030 |
0.1055186862 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 5 |
Hb_000656_120 |
0.1077542321 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 6 |
Hb_150745_010 |
0.1122151679 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 7 |
Hb_000953_180 |
0.1219745626 |
- |
- |
- |
| 8 |
Hb_008948_070 |
0.124870863 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003226_060 |
0.130181696 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_007101_250 |
0.1344129533 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 11 |
Hb_000397_170 |
0.1434143782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000771_040 |
0.14470291 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 13 |
Hb_006478_140 |
0.1458205381 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 14 |
Hb_096974_010 |
0.1561803735 |
- |
- |
PREDICTED: uncharacterized protein LOC105123384 isoform X1 [Populus euphratica] |
| 15 |
Hb_008115_030 |
0.1568944729 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 16 |
Hb_100395_010 |
0.1584864599 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001581_040 |
0.15901235 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000192_030 |
0.1643266445 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 19 |
Hb_005599_010 |
0.1652514101 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_000574_180 |
0.1659942894 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |