| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
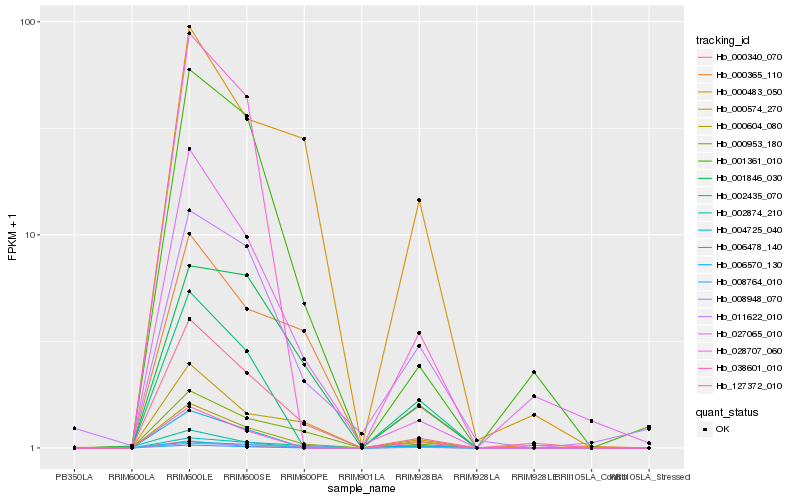
Hb_000604_080 |
0.0 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 2 |
Hb_006478_140 |
0.0870354801 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 3 |
Hb_000340_070 |
0.1223115748 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 4 |
Hb_127372_010 |
0.1320211361 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 5 |
Hb_038601_010 |
0.1480057942 |
- |
- |
PREDICTED: uncharacterized protein LOC102606280 [Solanum tuberosum] |
| 6 |
Hb_002435_070 |
0.1493986905 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 7 |
Hb_004725_040 |
0.1583629897 |
- |
- |
- |
| 8 |
Hb_011622_010 |
0.1602495537 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 9 |
Hb_000574_270 |
0.1607678978 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 10 |
Hb_000953_180 |
0.1618911486 |
- |
- |
- |
| 11 |
Hb_002874_210 |
0.1668357019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006570_130 |
0.1672052795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001361_010 |
0.1753623885 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_000483_050 |
0.1789737861 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 15 |
Hb_008948_070 |
0.1862053729 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000365_110 |
0.1870542071 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 17 |
Hb_001846_030 |
0.1911956025 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 18 |
Hb_008764_010 |
0.1936484445 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_027065_010 |
0.1940984833 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 20 |
Hb_028707_060 |
0.1950808175 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC101308915 [Fragaria vesca subsp. vesca] |