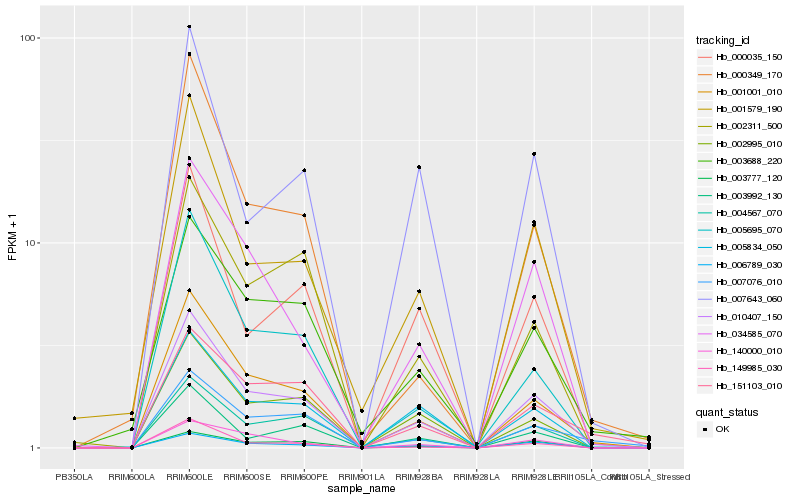
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_150 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 2 |
Hb_001001_010 |
0.0596910712 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 3 |
Hb_006789_030 |
0.1064521313 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 4 |
Hb_004567_070 |
0.1142420567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005695_070 |
0.1199423246 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 6 |
Hb_005834_050 |
0.1205225698 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 7 |
Hb_002995_010 |
0.1205409142 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 8 |
Hb_003777_120 |
0.128475777 |
- |
- |
- |
| 9 |
Hb_001579_190 |
0.1285818978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000035_150 |
0.1292734906 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 11 |
Hb_149985_030 |
0.1336999991 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_007076_010 |
0.1337868043 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_034585_070 |
0.133882422 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 14 |
Hb_003688_220 |
0.1353635999 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_003992_130 |
0.1354630115 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002311_500 |
0.1392412168 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 17 |
Hb_151103_010 |
0.1408997539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000349_170 |
0.1414623338 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 19 |
Hb_140000_010 |
0.1440225603 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_007643_060 |
0.1450020035 |
- |
- |
- |