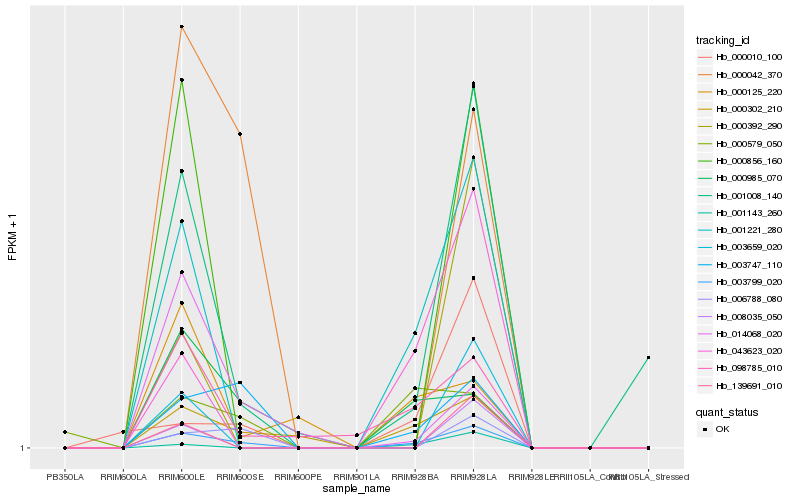
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003799_020 |
0.0 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 2 |
Hb_003747_110 |
0.2210951022 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 3 |
Hb_000302_210 |
0.224494025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001221_280 |
0.2538374908 |
- |
- |
- |
| 5 |
Hb_001008_140 |
0.2624529367 |
- |
- |
- |
| 6 |
Hb_098785_010 |
0.2673264747 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 7 |
Hb_006788_080 |
0.2908422568 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_043623_020 |
0.2935763526 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 9 |
Hb_000985_070 |
0.294255983 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 10 |
Hb_000042_370 |
0.3037027814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000010_100 |
0.3039404373 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 12 |
Hb_001143_260 |
0.3081096364 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 13 |
Hb_000579_050 |
0.3129467155 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 14 |
Hb_014068_020 |
0.3453820332 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 15 |
Hb_003659_020 |
0.3456964322 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 16 |
Hb_008035_050 |
0.3465504048 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 17 |
Hb_000125_220 |
0.3470741159 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 13 [Jatropha curcas] |
| 18 |
Hb_139691_010 |
0.3483876486 |
- |
- |
- |
| 19 |
Hb_000856_160 |
0.3503366619 |
- |
- |
- |
| 20 |
Hb_000392_290 |
0.3610690862 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |