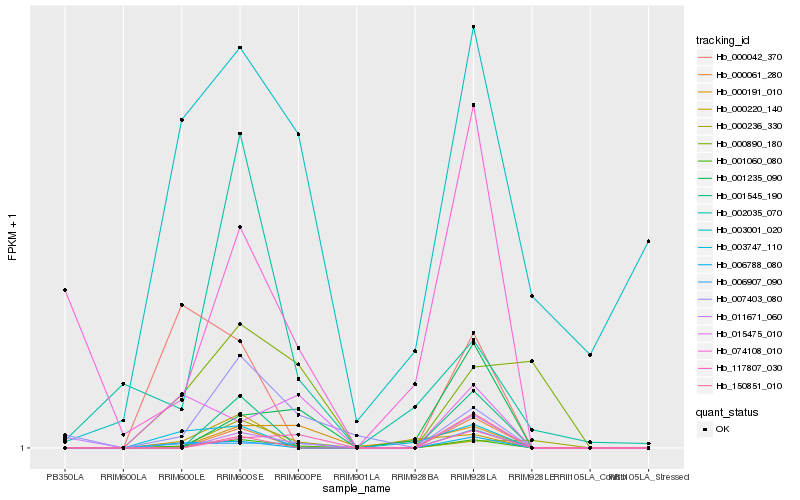
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001060_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 2 |
Hb_006907_090 |
0.257232971 |
- |
- |
- |
| 3 |
Hb_006788_080 |
0.2777720536 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 4 |
Hb_007403_080 |
0.3107843555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001545_190 |
0.3124162537 |
- |
- |
hypothetical protein VITISV_012031 [Vitis vinifera] |
| 6 |
Hb_000061_280 |
0.3252371715 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 7 |
Hb_117807_030 |
0.3327810406 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 8 |
Hb_003747_110 |
0.3433966443 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 9 |
Hb_074108_010 |
0.3585702413 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_000191_010 |
0.3616052127 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_015475_010 |
0.3626245991 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 12 |
Hb_001235_090 |
0.3634855029 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 13 |
Hb_003001_020 |
0.3692986619 |
- |
- |
hypothetical protein JCGZ_17508 [Jatropha curcas] |
| 14 |
Hb_000042_370 |
0.3754942257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000220_140 |
0.3795717351 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 16 |
Hb_150851_010 |
0.3805788988 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000236_330 |
0.3828378081 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_011671_060 |
0.3855330223 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 19 |
Hb_002035_070 |
0.3856812997 |
- |
- |
hypothetical protein CICLE_v10033569mg [Citrus clementina] |
| 20 |
Hb_000890_180 |
0.3870437336 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |