| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
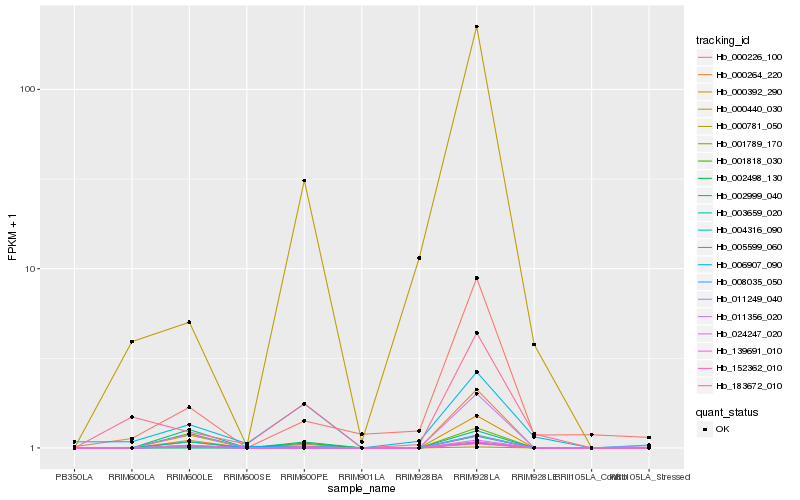
Hb_152362_010 |
0.0 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 2 |
Hb_000264_220 |
0.2208428252 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 3 |
Hb_006907_090 |
0.2784606097 |
- |
- |
- |
| 4 |
Hb_002999_040 |
0.282448116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000440_030 |
0.2927094815 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 6 |
Hb_002498_130 |
0.2979120878 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |
| 7 |
Hb_000781_050 |
0.2979567206 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_011356_020 |
0.2988189285 |
- |
- |
- |
| 9 |
Hb_004316_090 |
0.2993685702 |
- |
- |
hypothetical protein JCGZ_21197 [Jatropha curcas] |
| 10 |
Hb_001789_170 |
0.2998509375 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 11 |
Hb_005599_060 |
0.305455253 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001818_030 |
0.3055262104 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 13 |
Hb_000392_290 |
0.3073053008 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_024247_020 |
0.3111787131 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 15 |
Hb_000226_100 |
0.3126108711 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_183672_010 |
0.3142870664 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 17 |
Hb_011249_040 |
0.3187976753 |
- |
- |
PREDICTED: acetylajmalan esterase-like [Populus euphratica] |
| 18 |
Hb_139691_010 |
0.3219989854 |
- |
- |
- |
| 19 |
Hb_008035_050 |
0.3256352314 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 20 |
Hb_003659_020 |
0.327594714 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |