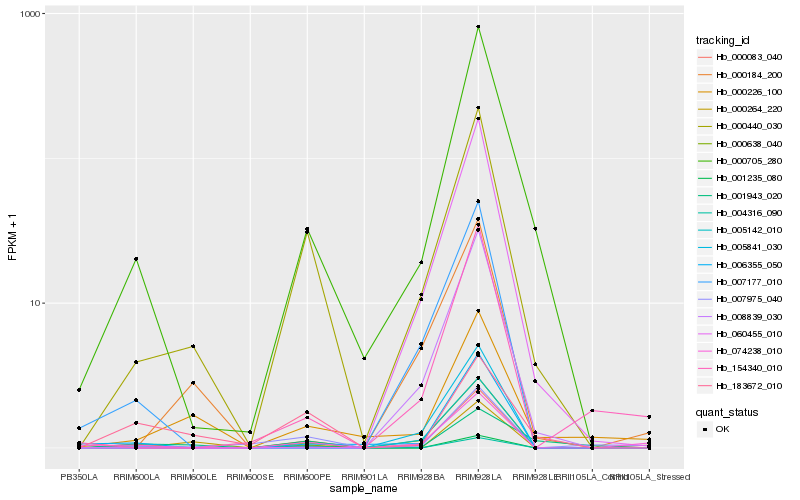
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000440_030 |
0.0 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 2 |
Hb_000705_280 |
0.1805473606 |
- |
- |
PREDICTED: uncharacterized protein LOC104601116 [Nelumbo nucifera] |
| 3 |
Hb_000638_040 |
0.1866193296 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-7-like [Glycine max] |
| 4 |
Hb_004316_090 |
0.1954692714 |
- |
- |
hypothetical protein JCGZ_21197 [Jatropha curcas] |
| 5 |
Hb_007975_040 |
0.2165166054 |
- |
- |
PREDICTED: uncharacterized protein LOC104610918 [Nelumbo nucifera] |
| 6 |
Hb_000226_100 |
0.2308991362 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 7 |
Hb_074238_010 |
0.2394754928 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 8 |
Hb_183672_010 |
0.2447528954 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 9 |
Hb_006355_050 |
0.2476656276 |
- |
- |
- |
| 10 |
Hb_000264_220 |
0.2488458009 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 11 |
Hb_000083_040 |
0.2625923312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_154340_010 |
0.2632145135 |
- |
- |
truncated nitrate transporter [Fragaria vesca] |
| 13 |
Hb_001943_020 |
0.2636139215 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 14 |
Hb_000184_200 |
0.2637318696 |
- |
- |
- |
| 15 |
Hb_001235_080 |
0.2650412809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_060455_010 |
0.2673486761 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 17 |
Hb_008839_030 |
0.267878488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005142_010 |
0.2791149449 |
- |
- |
- |
| 19 |
Hb_007177_010 |
0.2794675905 |
- |
- |
- |
| 20 |
Hb_005841_030 |
0.2795768151 |
- |
- |
- |