| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
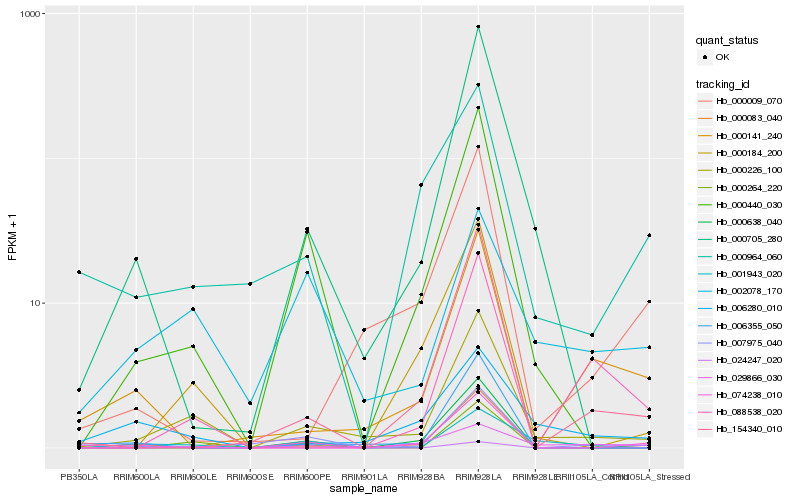
Hb_000226_100 |
0.0 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_000440_030 |
0.2308991362 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 3 |
Hb_000264_220 |
0.2385066075 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 4 |
Hb_000705_280 |
0.2422426079 |
- |
- |
PREDICTED: uncharacterized protein LOC104601116 [Nelumbo nucifera] |
| 5 |
Hb_001943_020 |
0.2506600211 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 6 |
Hb_000184_200 |
0.2509901203 |
- |
- |
- |
| 7 |
Hb_007975_040 |
0.2517591363 |
- |
- |
PREDICTED: uncharacterized protein LOC104610918 [Nelumbo nucifera] |
| 8 |
Hb_154340_010 |
0.2611856303 |
- |
- |
truncated nitrate transporter [Fragaria vesca] |
| 9 |
Hb_000083_040 |
0.2643320429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006280_010 |
0.2700014994 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006355_050 |
0.2715054902 |
- |
- |
- |
| 12 |
Hb_000009_070 |
0.2753094882 |
- |
- |
PREDICTED: globulin-1 S allele [Jatropha curcas] |
| 13 |
Hb_074238_010 |
0.2831947859 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 14 |
Hb_024247_020 |
0.2837540232 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 15 |
Hb_000638_040 |
0.2908131548 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-7-like [Glycine max] |
| 16 |
Hb_002078_170 |
0.2911032327 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 17 |
Hb_000141_240 |
0.2919879157 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_029866_030 |
0.2923336331 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 19 |
Hb_000964_060 |
0.2941664317 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 20 |
Hb_088538_020 |
0.297316374 |
- |
- |
hypothetical protein JCGZ_12178 [Jatropha curcas] |