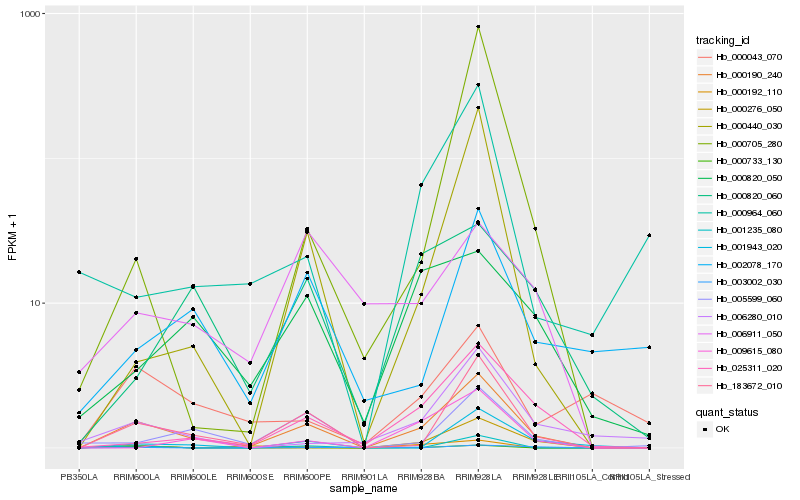
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_240 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_183672_010 |
0.1939341647 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 3 |
Hb_025311_020 |
0.2635294976 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 4 |
Hb_001943_020 |
0.2659705985 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 5 |
Hb_000440_030 |
0.2964091807 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 6 |
Hb_005599_060 |
0.3001262687 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006280_010 |
0.3022974435 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_009615_080 |
0.3149014719 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 9 |
Hb_000820_050 |
0.3219944284 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 10 |
Hb_002078_170 |
0.3226763283 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_000820_060 |
0.3234910125 |
- |
- |
- |
| 12 |
Hb_000043_070 |
0.3275268897 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 13 |
Hb_001235_080 |
0.3282730006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000964_060 |
0.3360128321 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 15 |
Hb_000192_110 |
0.3382842556 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 16 |
Hb_003002_030 |
0.338296791 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_000276_050 |
0.3395810933 |
- |
- |
- |
| 18 |
Hb_000733_130 |
0.3415260135 |
- |
- |
PREDICTED: cucumisin [Vitis vinifera] |
| 19 |
Hb_006911_050 |
0.3429217237 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 20 |
Hb_000705_280 |
0.3435013439 |
- |
- |
PREDICTED: uncharacterized protein LOC104601116 [Nelumbo nucifera] |