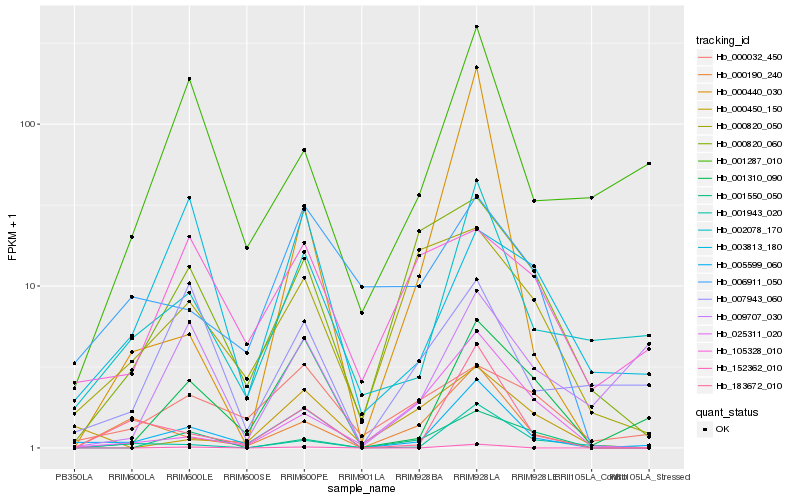
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005599_060 |
0.0 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002078_170 |
0.2092880504 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 3 |
Hb_001310_090 |
0.2232864369 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 4 |
Hb_183672_010 |
0.2497426498 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 5 |
Hb_000190_240 |
0.3001262687 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000032_450 |
0.3032973212 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 7 |
Hb_001287_010 |
0.3039566572 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 8 |
Hb_152362_010 |
0.305455253 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 9 |
Hb_006911_050 |
0.3055319815 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 10 |
Hb_007943_060 |
0.3137651535 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 11 |
Hb_000450_150 |
0.3146658507 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001943_020 |
0.3170061773 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 13 |
Hb_001550_050 |
0.3300624805 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Eucalyptus grandis] |
| 14 |
Hb_000820_050 |
0.3306103071 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 15 |
Hb_000820_060 |
0.3322409363 |
- |
- |
- |
| 16 |
Hb_000440_030 |
0.3322519242 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 17 |
Hb_009707_030 |
0.3367582639 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 18 |
Hb_025311_020 |
0.340061744 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 19 |
Hb_003813_180 |
0.3411541709 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 20 |
Hb_105328_010 |
0.3430821965 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |