| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
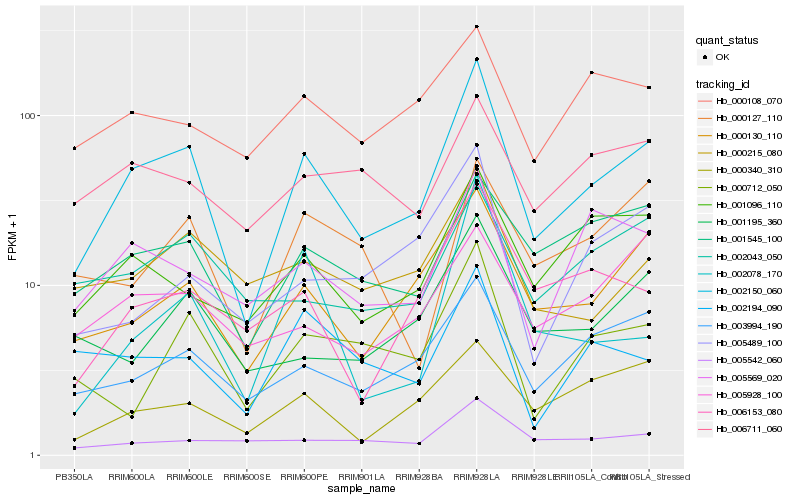
Hb_002150_060 |
0.0 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 2 |
Hb_000130_110 |
0.1533560667 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 3 |
Hb_003994_190 |
0.1701539299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001545_100 |
0.1828393216 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 5 |
Hb_001096_110 |
0.1881094011 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 6 |
Hb_006153_080 |
0.1886624649 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 7 |
Hb_000712_050 |
0.1886645342 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_000340_310 |
0.188982134 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 9 |
Hb_005569_020 |
0.192174488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001195_360 |
0.1947952671 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 11 |
Hb_000215_080 |
0.195536369 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005542_060 |
0.2011310444 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002043_050 |
0.2037824 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 14 |
Hb_005928_100 |
0.2053511068 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 15 |
Hb_002194_090 |
0.2059061433 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000127_110 |
0.2073556892 |
- |
- |
- |
| 17 |
Hb_005489_100 |
0.2076761157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000108_070 |
0.2089073683 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 19 |
Hb_006711_060 |
0.2096468705 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 20 |
Hb_002078_170 |
0.2104678214 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |