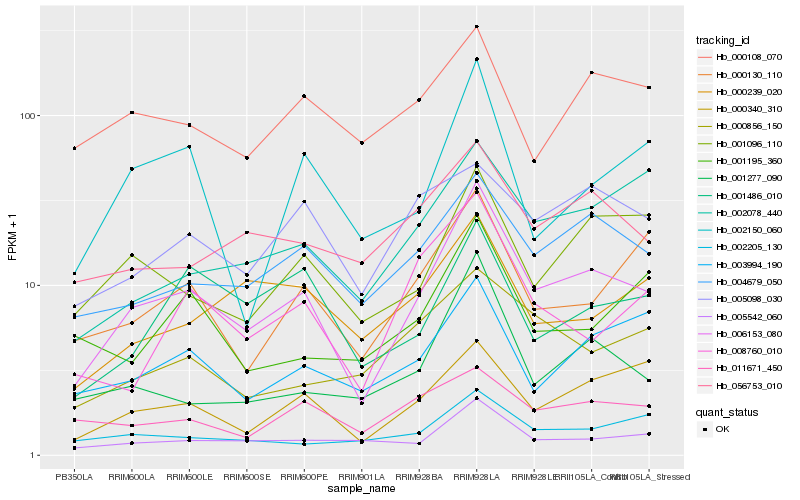
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_080 |
0.0 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 2 |
Hb_004679_050 |
0.1574163461 |
- |
- |
galactono-1,4-lactone dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_005542_060 |
0.1660007066 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000340_310 |
0.1672974557 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 5 |
Hb_001277_090 |
0.169130835 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 6 |
Hb_001096_110 |
0.1736576773 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 7 |
Hb_000130_110 |
0.1798557185 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 8 |
Hb_056753_010 |
0.1807633423 |
- |
- |
PREDICTED: L-galactono-1,4-lactone dehydrogenase, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_008760_010 |
0.1833681272 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 10 |
Hb_003994_190 |
0.1851372577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002150_060 |
0.1886624649 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 12 |
Hb_002078_440 |
0.1899745139 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002205_130 |
0.1910347144 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 14 |
Hb_001486_010 |
0.1932004197 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 15 |
Hb_000239_020 |
0.1949267921 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 16 |
Hb_000108_070 |
0.1949659413 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 17 |
Hb_000856_150 |
0.1961118561 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 18 |
Hb_005098_030 |
0.1968487846 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011671_450 |
0.1987965504 |
- |
- |
- |
| 20 |
Hb_001195_360 |
0.2019469156 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |