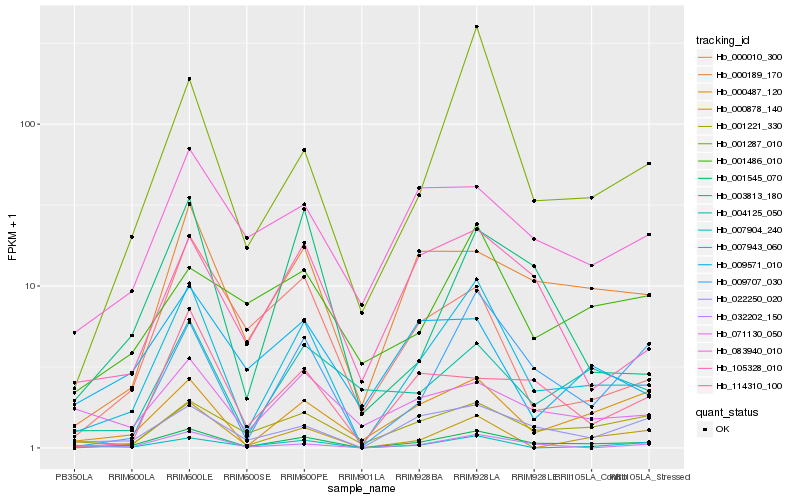
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_060 |
0.0 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 2 |
Hb_001545_070 |
0.096891162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001287_010 |
0.1856947794 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 4 |
Hb_009707_030 |
0.2057087606 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 5 |
Hb_032202_150 |
0.2163596048 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 6 |
Hb_000878_140 |
0.2179817995 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_000487_120 |
0.2228445728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000189_170 |
0.229296103 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_001221_330 |
0.2331455937 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 10 |
Hb_105328_010 |
0.2333710752 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 11 |
Hb_003813_180 |
0.2371836393 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_022250_020 |
0.2384094323 |
- |
- |
PREDICTED: uncharacterized protein LOC105648703 [Jatropha curcas] |
| 13 |
Hb_004125_050 |
0.2402136459 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 14 |
Hb_007904_240 |
0.2418619344 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 15 |
Hb_000010_300 |
0.245857805 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_071130_050 |
0.2594196886 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 17 |
Hb_009571_010 |
0.2622075035 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 18 |
Hb_083940_010 |
0.2639263613 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 19 |
Hb_001486_010 |
0.2641091626 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 20 |
Hb_114310_100 |
0.2676426513 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |