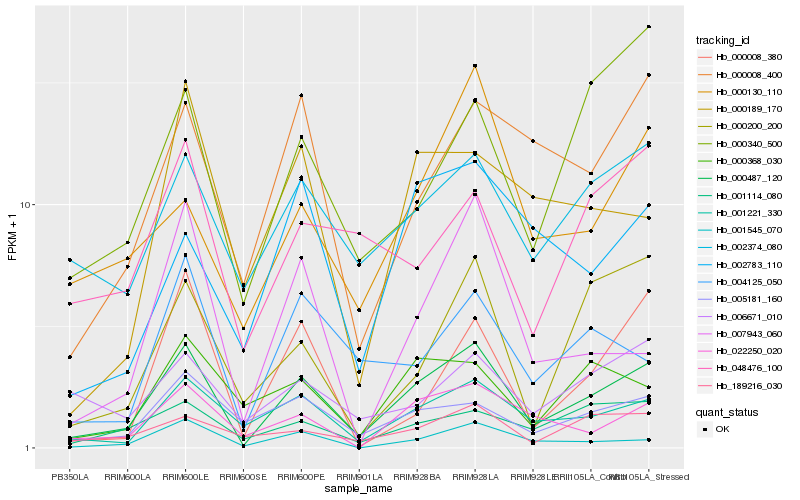
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001221_330 |
0.1665745915 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 3 |
Hb_001545_070 |
0.1921578813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005181_160 |
0.1931553838 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 5 |
Hb_022250_020 |
0.195345958 |
- |
- |
PREDICTED: uncharacterized protein LOC105648703 [Jatropha curcas] |
| 6 |
Hb_002374_080 |
0.1984895973 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001114_080 |
0.1986347234 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 8 |
Hb_000008_400 |
0.202518263 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 9 |
Hb_189216_030 |
0.2059734807 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 10 |
Hb_004125_050 |
0.2108793128 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 11 |
Hb_000368_030 |
0.2127461685 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 12 |
Hb_000200_200 |
0.2134955355 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000130_110 |
0.2171981956 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 14 |
Hb_002783_110 |
0.2203604935 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 15 |
Hb_006671_010 |
0.221224481 |
- |
- |
PREDICTED: 5' exonuclease Apollo isoform X1 [Jatropha curcas] |
| 16 |
Hb_007943_060 |
0.2228445728 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 17 |
Hb_000340_500 |
0.2228477284 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 18 |
Hb_000008_380 |
0.2235282988 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000189_170 |
0.2242015452 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_048476_100 |
0.2246441331 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |