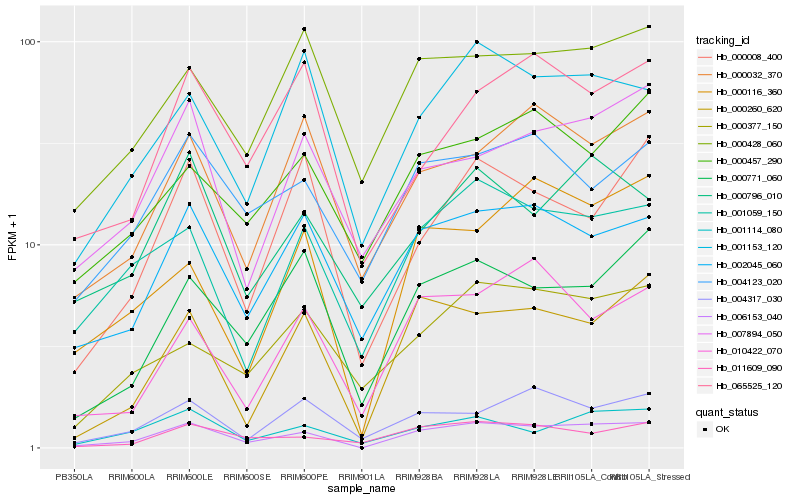
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_040 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 2 |
Hb_002045_060 |
0.1438862031 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000260_620 |
0.1451552482 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |
| 4 |
Hb_001059_150 |
0.149172192 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 5 |
Hb_001114_080 |
0.1534039245 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 6 |
Hb_000771_060 |
0.1579154527 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 7 |
Hb_001153_120 |
0.1620200431 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 8 |
Hb_004123_020 |
0.1637470225 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000796_010 |
0.163923445 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 10 |
Hb_000428_060 |
0.1651837963 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_007894_050 |
0.1656672558 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000116_360 |
0.1657463552 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 13 |
Hb_011609_090 |
0.166307806 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_004317_030 |
0.166544152 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 15 |
Hb_000032_370 |
0.1672457332 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 16 |
Hb_000008_400 |
0.1708972922 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 17 |
Hb_010422_070 |
0.1733421021 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 18 |
Hb_000377_150 |
0.1744086919 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 19 |
Hb_065525_120 |
0.1757561655 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 20 |
Hb_000457_290 |
0.1760700664 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |