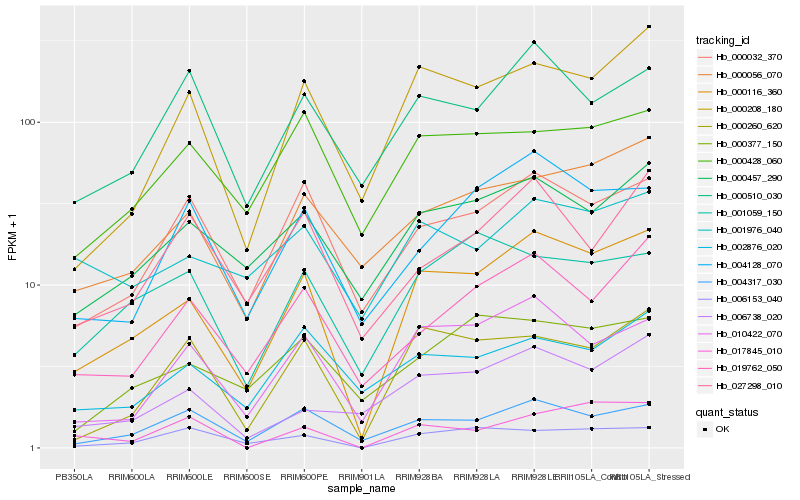
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_360 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 2 |
Hb_010422_070 |
0.1354358979 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |
| 3 |
Hb_000208_180 |
0.1396599986 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 4 |
Hb_000457_290 |
0.1463038594 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 5 |
Hb_004317_030 |
0.1498601334 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 6 |
Hb_000056_070 |
0.1501051184 |
- |
- |
- |
| 7 |
Hb_000260_620 |
0.1502075172 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |
| 8 |
Hb_019762_050 |
0.1528039349 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 9 |
Hb_000032_370 |
0.1531561429 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 10 |
Hb_004128_070 |
0.1626254274 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006153_040 |
0.1657463552 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 12 |
Hb_006738_020 |
0.1660268468 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 13 |
Hb_000377_150 |
0.1712222638 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 14 |
Hb_000510_030 |
0.1714943926 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 15 |
Hb_000428_060 |
0.172293309 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001059_150 |
0.174374948 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 17 |
Hb_027298_010 |
0.1766448698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002876_020 |
0.1767298553 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X2 [Jatropha curcas] |
| 19 |
Hb_017845_010 |
0.1777468863 |
- |
- |
- |
| 20 |
Hb_001976_040 |
0.1788165121 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |