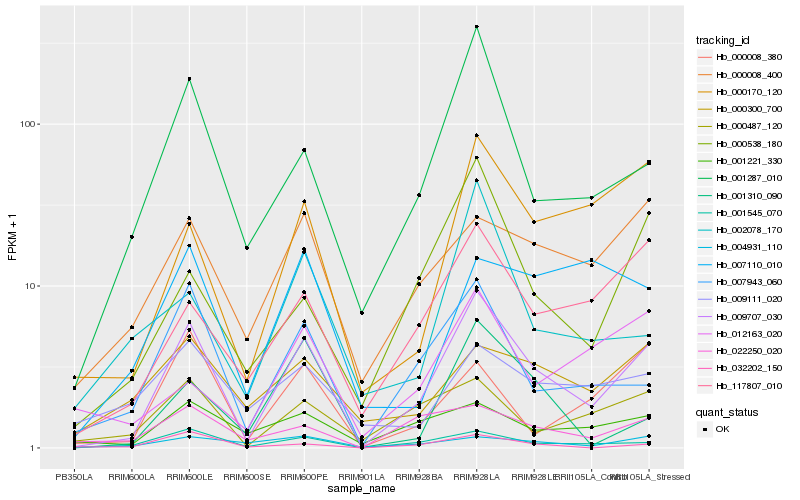
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009707_030 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 2 |
Hb_001545_070 |
0.1955774713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001287_010 |
0.2052929565 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 4 |
Hb_007943_060 |
0.2057087606 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 5 |
Hb_117807_010 |
0.2243953395 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 6 |
Hb_000170_120 |
0.2281108796 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000008_400 |
0.2388108468 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 8 |
Hb_000487_120 |
0.2407881931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000538_180 |
0.2422100533 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 10 |
Hb_001221_330 |
0.2467078657 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 11 |
Hb_022250_020 |
0.2510603651 |
- |
- |
PREDICTED: uncharacterized protein LOC105648703 [Jatropha curcas] |
| 12 |
Hb_001310_090 |
0.2551165347 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 13 |
Hb_032202_150 |
0.2562245532 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 14 |
Hb_004931_110 |
0.2565829266 |
- |
- |
inorganic phosphate transporter, putative [Ricinus communis] |
| 15 |
Hb_000008_380 |
0.256631716 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000300_700 |
0.2611346792 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3 [Jatropha curcas] |
| 17 |
Hb_002078_170 |
0.2653730996 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 18 |
Hb_009111_020 |
0.2691117444 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 19 |
Hb_012163_020 |
0.2709978578 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 20 |
Hb_007110_010 |
0.2773482298 |
- |
- |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |