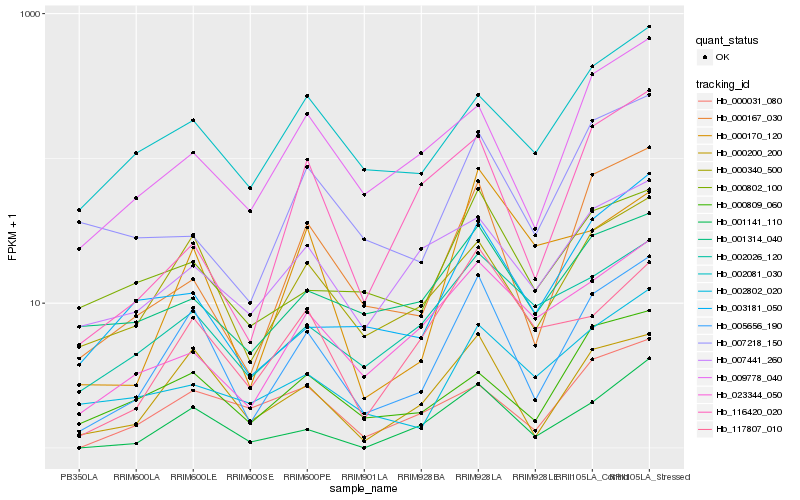
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_190 |
0.0 |
- |
- |
proteasome subunit beta type 6,9, putative [Ricinus communis] |
| 2 |
Hb_000200_200 |
0.1262080966 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000340_500 |
0.1488433707 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 4 |
Hb_001141_110 |
0.156447789 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 5 |
Hb_000167_030 |
0.1610446094 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009778_040 |
0.1952922939 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 7 |
Hb_003181_050 |
0.1983313438 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 8 |
Hb_000809_060 |
0.2011731169 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 9 |
Hb_002026_120 |
0.2013797499 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 10 |
Hb_000031_080 |
0.2015419513 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 11 |
Hb_002081_030 |
0.2025682029 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_117807_010 |
0.2040749894 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 13 |
Hb_002802_020 |
0.206144907 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 14 |
Hb_023344_050 |
0.2082964174 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 15 |
Hb_000802_100 |
0.208474608 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 16 |
Hb_116420_020 |
0.2093465198 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 17 |
Hb_000170_120 |
0.2098004308 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_007441_260 |
0.2138242701 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_007218_150 |
0.2139782353 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 20 |
Hb_001314_040 |
0.2143925378 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |