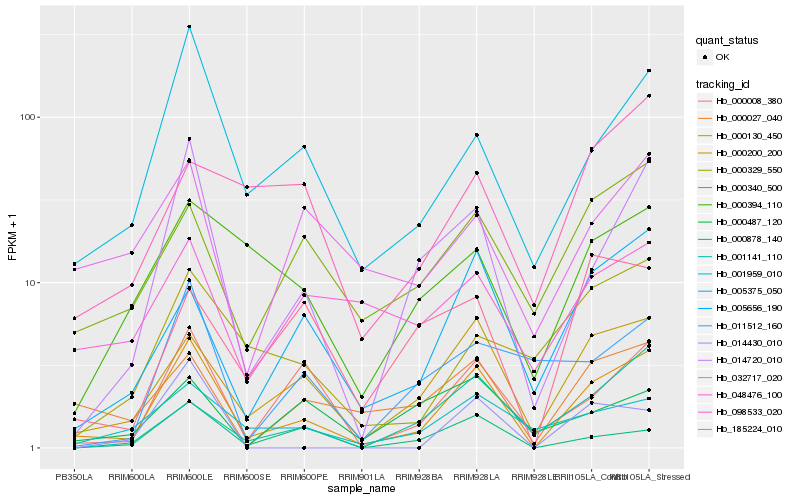
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_450 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000200_200 |
0.179740706 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 3 |
Hb_014720_010 |
0.1811509416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005375_050 |
0.2049796595 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 5 |
Hb_000008_380 |
0.2131209177 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_005656_190 |
0.2300458537 |
- |
- |
proteasome subunit beta type 6,9, putative [Ricinus communis] |
| 7 |
Hb_000027_040 |
0.2316759759 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |
| 8 |
Hb_001959_010 |
0.2393164059 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 9 |
Hb_000340_500 |
0.2463920414 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 10 |
Hb_000878_140 |
0.250745484 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 11 |
Hb_000329_550 |
0.2649906673 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 12 |
Hb_000394_110 |
0.2675458329 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_032717_020 |
0.2692659736 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011512_160 |
0.2711480567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_185224_010 |
0.271789283 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_001141_110 |
0.2744047852 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 17 |
Hb_048476_100 |
0.2764582436 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |
| 18 |
Hb_014430_010 |
0.278875003 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_098533_020 |
0.2833500595 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 20 |
Hb_000487_120 |
0.2837109968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |