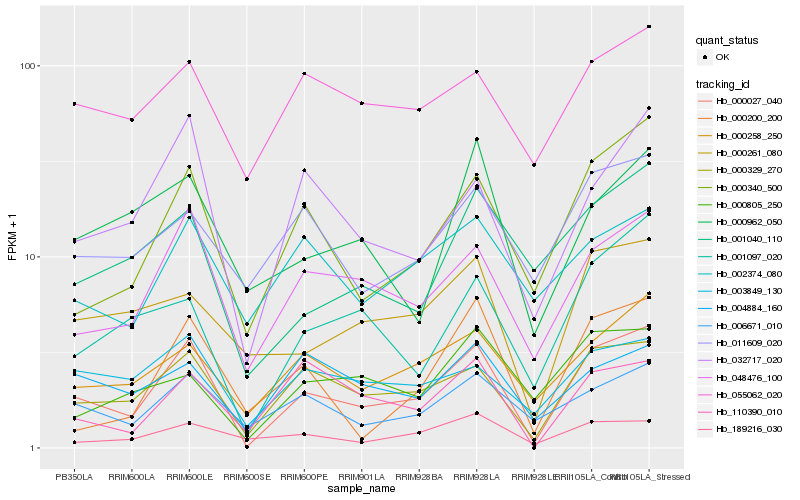
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |
| 2 |
Hb_000329_270 |
0.1385708476 |
- |
- |
PREDICTED: spindle and kinetochore-associated protein 1 homolog [Jatropha curcas] |
| 3 |
Hb_048476_100 |
0.1603185589 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |
| 4 |
Hb_000340_500 |
0.1675395179 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 5 |
Hb_032717_020 |
0.170879401 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000258_250 |
0.1851843395 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 7 |
Hb_006671_010 |
0.1854896242 |
- |
- |
PREDICTED: 5' exonuclease Apollo isoform X1 [Jatropha curcas] |
| 8 |
Hb_001097_020 |
0.1909818515 |
- |
- |
hypothetical protein POPTR_0011s00490g [Populus trichocarpa] |
| 9 |
Hb_000261_080 |
0.1974734051 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 10 |
Hb_110390_010 |
0.2008480446 |
- |
- |
hypothetical protein JCGZ_19741 [Jatropha curcas] |
| 11 |
Hb_189216_030 |
0.2009909477 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 12 |
Hb_003849_130 |
0.2011316743 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 13 |
Hb_001040_110 |
0.2014730072 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 14 |
Hb_000200_200 |
0.2025148556 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011609_020 |
0.2119485333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000805_250 |
0.2122235434 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 17 |
Hb_004884_160 |
0.2132003503 |
- |
- |
PREDICTED: uncharacterized protein LOC105648105 [Jatropha curcas] |
| 18 |
Hb_055062_020 |
0.2141626225 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 19 |
Hb_000962_050 |
0.215119395 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 20 |
Hb_002374_080 |
0.2159035737 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |