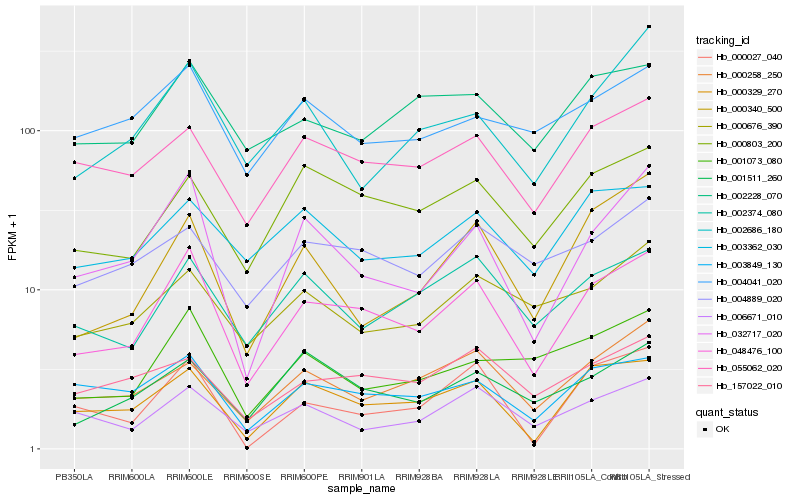
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_100 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |
| 2 |
Hb_000329_270 |
0.1174141984 |
- |
- |
PREDICTED: spindle and kinetochore-associated protein 1 homolog [Jatropha curcas] |
| 3 |
Hb_032717_020 |
0.1174986796 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003849_130 |
0.1331129688 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 5 |
Hb_000803_200 |
0.1336929132 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 6 |
Hb_055062_020 |
0.1359154811 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_002374_080 |
0.1383894209 |
- |
- |
PREDICTED: chloroplast processing peptidase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_006671_010 |
0.1414863116 |
- |
- |
PREDICTED: 5' exonuclease Apollo isoform X1 [Jatropha curcas] |
| 9 |
Hb_000258_250 |
0.143385306 |
- |
- |
Origin recognition complex subunit, putative [Ricinus communis] |
| 10 |
Hb_002228_070 |
0.1447609752 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 11 |
Hb_001511_260 |
0.1462982346 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 12 |
Hb_157022_010 |
0.1464840334 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 13 |
Hb_001073_080 |
0.147295675 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003362_030 |
0.1502070768 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 15 |
Hb_004889_020 |
0.1539733456 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 16 |
Hb_000676_390 |
0.1543616424 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000340_500 |
0.1550203835 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 18 |
Hb_004041_020 |
0.156446431 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 19 |
Hb_002686_180 |
0.1595819987 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 20 |
Hb_000027_040 |
0.1603185589 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |