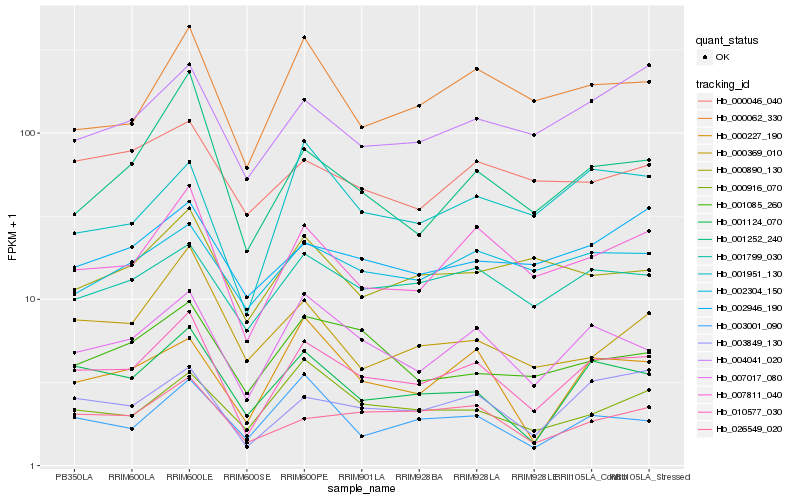
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010577_030 |
0.0 |
- |
- |
PREDICTED: putative cyclin-B3-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007017_080 |
0.0920648833 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_007811_040 |
0.0924504063 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 4 |
Hb_003849_130 |
0.1077360869 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 5 |
Hb_001124_070 |
0.1089373587 |
- |
- |
Serine/threonine-protein kinase ULK4 [Gossypium arboreum] |
| 6 |
Hb_026549_020 |
0.1189456072 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001799_030 |
0.1193426205 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 8 |
Hb_000062_330 |
0.1299043496 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_000227_190 |
0.1344035548 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004041_020 |
0.1355650321 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 11 |
Hb_001252_240 |
0.1363403231 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 12 |
Hb_001085_260 |
0.1368497641 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 13 |
Hb_000916_070 |
0.1370501126 |
- |
- |
PREDICTED: sister chromatid cohesion protein DCC1-like [Jatropha curcas] |
| 14 |
Hb_003001_090 |
0.1374674215 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 15 |
Hb_002304_150 |
0.1374860334 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001951_130 |
0.1384525831 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 17 |
Hb_002946_190 |
0.1417733768 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000046_040 |
0.1431617946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000369_010 |
0.1444599392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000890_130 |
0.1450809398 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |