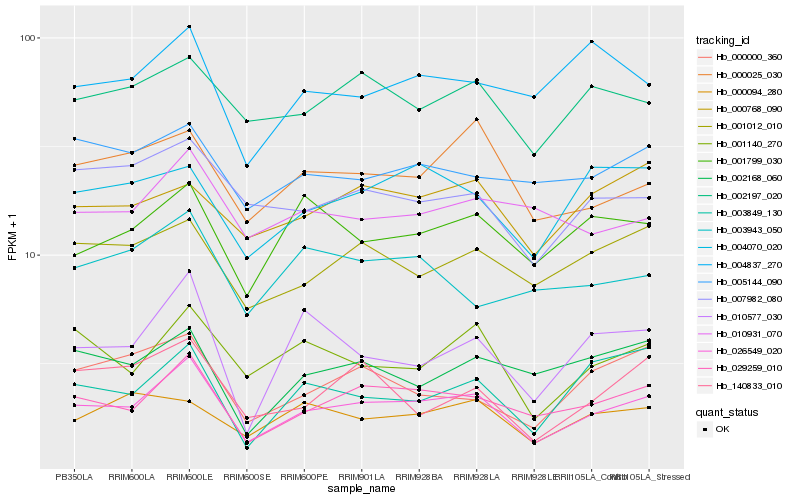
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026549_020 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_029259_010 |
0.080958584 |
- |
- |
hypothetical protein JCGZ_20186 [Jatropha curcas] |
| 3 |
Hb_001140_270 |
0.1136068142 |
- |
- |
hypothetical protein EUGRSUZ_E00166 [Eucalyptus grandis] |
| 4 |
Hb_010577_030 |
0.1189456072 |
- |
- |
PREDICTED: putative cyclin-B3-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000025_030 |
0.1222315596 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001012_010 |
0.1237182528 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 7 |
Hb_007982_080 |
0.1244388877 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 8 |
Hb_001799_030 |
0.1262247683 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 9 |
Hb_002197_020 |
0.1264857064 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 10 |
Hb_004070_020 |
0.1289912603 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 11 |
Hb_000000_360 |
0.1293391125 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 12 |
Hb_003849_130 |
0.1305707991 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 13 |
Hb_002168_060 |
0.1317911077 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_140833_010 |
0.1324030129 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 15 |
Hb_005144_090 |
0.1353633642 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 16 |
Hb_010931_070 |
0.1355276892 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000768_090 |
0.1362610199 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Jatropha curcas] |
| 18 |
Hb_004837_270 |
0.137720745 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 19 |
Hb_000094_280 |
0.1383465253 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 20 |
Hb_003943_050 |
0.1384920853 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |