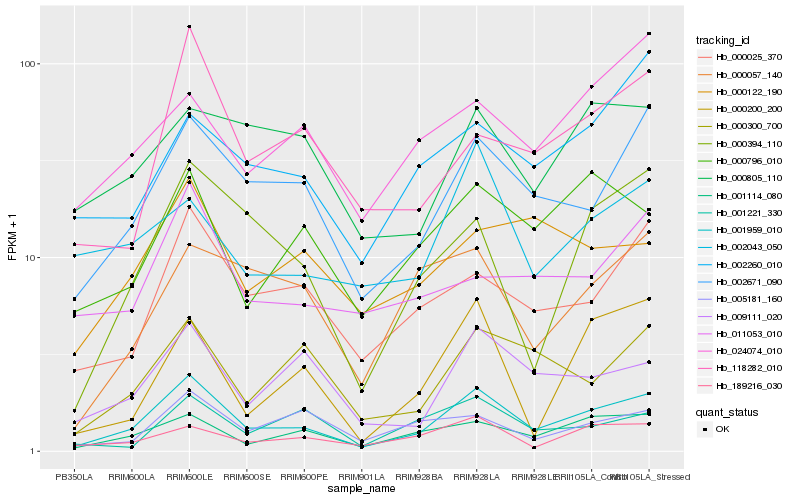
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_010 |
0.0 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 2 |
Hb_002671_090 |
0.1460834697 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 3 |
Hb_001114_080 |
0.1622660228 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 4 |
Hb_000025_370 |
0.1686641747 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000394_110 |
0.1724240679 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_009111_020 |
0.1777267332 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 7 |
Hb_000300_700 |
0.1825958313 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3 [Jatropha curcas] |
| 8 |
Hb_189216_030 |
0.1838327277 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 9 |
Hb_118282_010 |
0.1851188177 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 10 |
Hb_000796_010 |
0.1910711391 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 11 |
Hb_000200_200 |
0.1946099941 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000057_140 |
0.1987791209 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 13 |
Hb_005181_160 |
0.1990122219 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 14 |
Hb_000805_110 |
0.2063037631 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 15 |
Hb_001221_330 |
0.2065083158 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 16 |
Hb_000122_190 |
0.2072718686 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_011053_010 |
0.2076986425 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 18 |
Hb_002260_010 |
0.2090197961 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 19 |
Hb_002043_050 |
0.2092083672 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 20 |
Hb_024074_010 |
0.2096965855 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |