| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
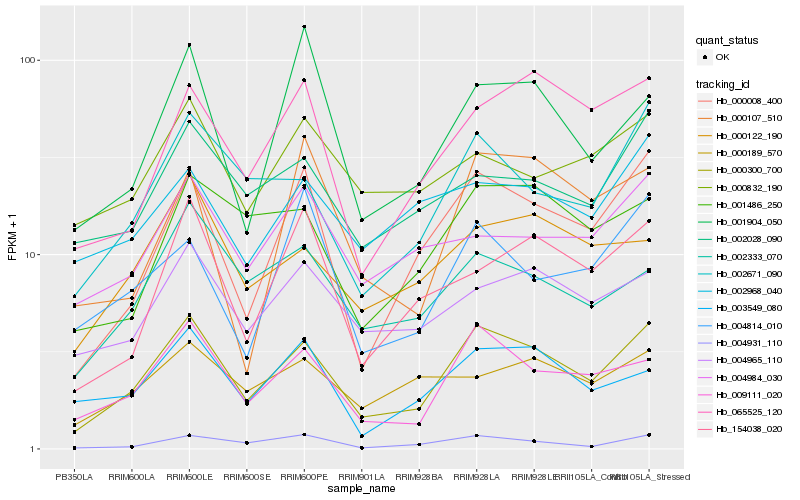
Hb_000300_700 |
0.0 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3 [Jatropha curcas] |
| 2 |
Hb_009111_020 |
0.1070128726 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 3 |
Hb_000008_400 |
0.1180208251 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 4 |
Hb_002671_090 |
0.1312474208 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 5 |
Hb_004931_110 |
0.1394745845 |
- |
- |
inorganic phosphate transporter, putative [Ricinus communis] |
| 6 |
Hb_065525_120 |
0.1490033657 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 7 |
Hb_004965_110 |
0.1513089222 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 8 |
Hb_002333_070 |
0.1515608701 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 9 |
Hb_001486_250 |
0.151795731 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 10 |
Hb_000122_190 |
0.1532061724 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003549_080 |
0.1536391723 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_154038_020 |
0.1548733456 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 13 |
Hb_002028_090 |
0.1593590792 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 14 |
Hb_001904_050 |
0.1636641553 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 15 |
Hb_004984_030 |
0.1659335769 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000189_570 |
0.165938747 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 17 |
Hb_002968_040 |
0.1711161241 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 18 |
Hb_000832_190 |
0.172504006 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 19 |
Hb_000107_510 |
0.1727206554 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 20 |
Hb_004814_010 |
0.1727732006 |
- |
- |
PREDICTED: uncharacterized protein LOC105633486 [Jatropha curcas] |