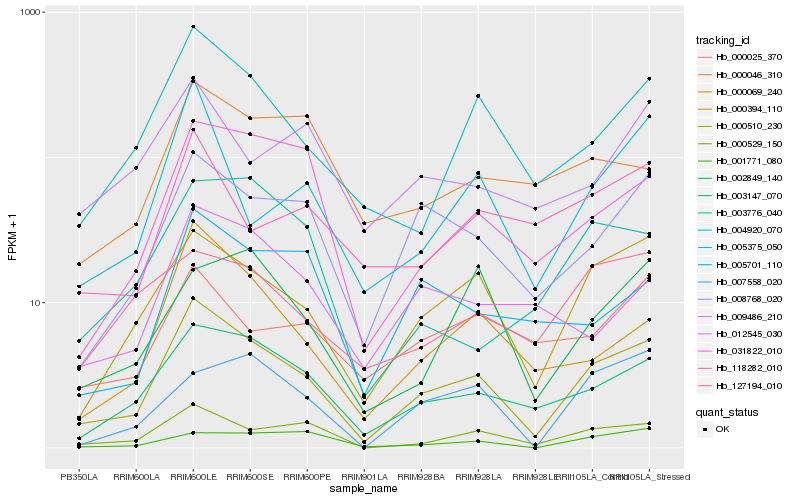
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_230 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 2 |
Hb_000394_110 |
0.1515774301 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 3 |
Hb_003147_070 |
0.1558810767 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 4 |
Hb_000529_150 |
0.1599533115 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_004920_070 |
0.171253008 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 6 |
Hb_008768_020 |
0.174475405 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 7 |
Hb_000069_240 |
0.1837919081 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 8 |
Hb_031822_010 |
0.1967562303 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 9 |
Hb_005375_050 |
0.2042089537 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 10 |
Hb_005701_110 |
0.2172750053 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 11 |
Hb_012545_030 |
0.2182435341 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 12 |
Hb_000025_370 |
0.2216084585 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 13 |
Hb_118282_010 |
0.2241227612 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 14 |
Hb_127194_010 |
0.2245087946 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 15 |
Hb_002849_140 |
0.2266831545 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 16 |
Hb_001771_080 |
0.2268845589 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 17 |
Hb_003776_040 |
0.2294394162 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 18 |
Hb_009486_210 |
0.2295144898 |
- |
- |
- |
| 19 |
Hb_007558_020 |
0.2299903745 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 20 |
Hb_000046_310 |
0.2302838636 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |