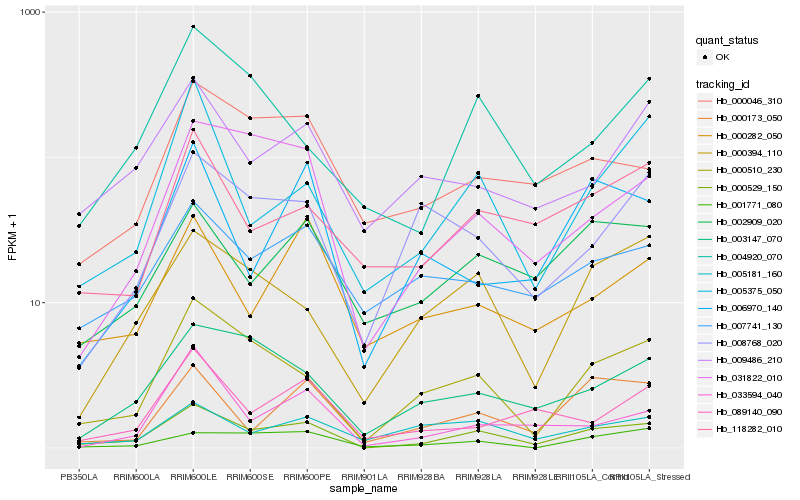
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_150 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000510_230 |
0.1599533115 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 3 |
Hb_031822_010 |
0.1877232346 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 4 |
Hb_000394_110 |
0.1948250539 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_003147_070 |
0.1979420519 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 6 |
Hb_002909_020 |
0.1995529655 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 7 |
Hb_118282_010 |
0.2042004882 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 8 |
Hb_005375_050 |
0.2047161288 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 9 |
Hb_000173_050 |
0.205135376 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 10 |
Hb_007741_130 |
0.2068157037 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 11 |
Hb_000046_310 |
0.2070243062 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 12 |
Hb_009486_210 |
0.2079108329 |
- |
- |
- |
| 13 |
Hb_004920_070 |
0.2104713714 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_005181_160 |
0.2114966678 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 15 |
Hb_000282_050 |
0.2143341396 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 16 |
Hb_001771_080 |
0.216013931 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 17 |
Hb_008768_020 |
0.2184643017 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 18 |
Hb_033594_040 |
0.219980343 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_006970_140 |
0.2200158206 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 20 |
Hb_089140_090 |
0.220712016 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |