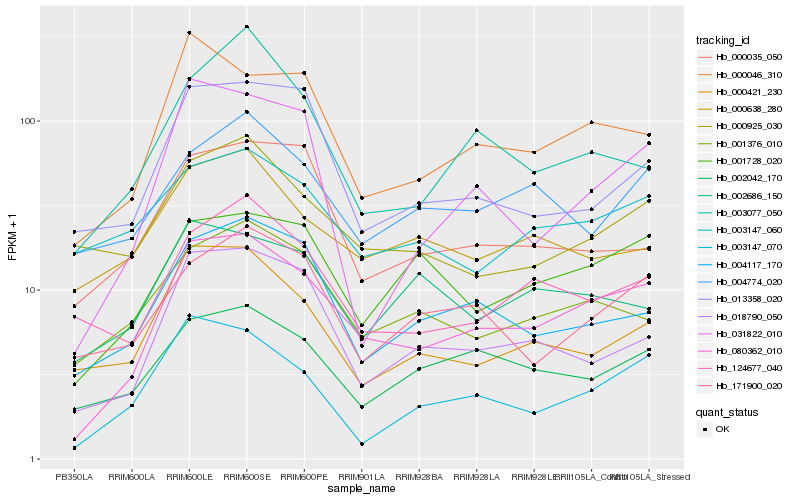
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080362_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 2 |
Hb_000046_310 |
0.1383730073 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 3 |
Hb_001728_020 |
0.1442846317 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 4 |
Hb_003147_070 |
0.1447042342 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 5 |
Hb_018790_050 |
0.1455133715 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 6 |
Hb_004117_170 |
0.1527547223 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 7 |
Hb_001376_010 |
0.1579878117 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 8 |
Hb_002042_170 |
0.1582908431 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 9 |
Hb_000421_230 |
0.1598532489 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 10 |
Hb_124677_040 |
0.1624893525 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 11 |
Hb_031822_010 |
0.162883748 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 12 |
Hb_171900_020 |
0.1654629976 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 13 |
Hb_013358_020 |
0.1686404843 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 14 |
Hb_004774_020 |
0.1696532257 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 15 |
Hb_000925_030 |
0.171560731 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_003077_050 |
0.1722161853 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 17 |
Hb_000638_280 |
0.1735439557 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_003147_060 |
0.1740002164 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 19 |
Hb_000035_050 |
0.1747201674 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 20 |
Hb_002686_150 |
0.1751524074 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |