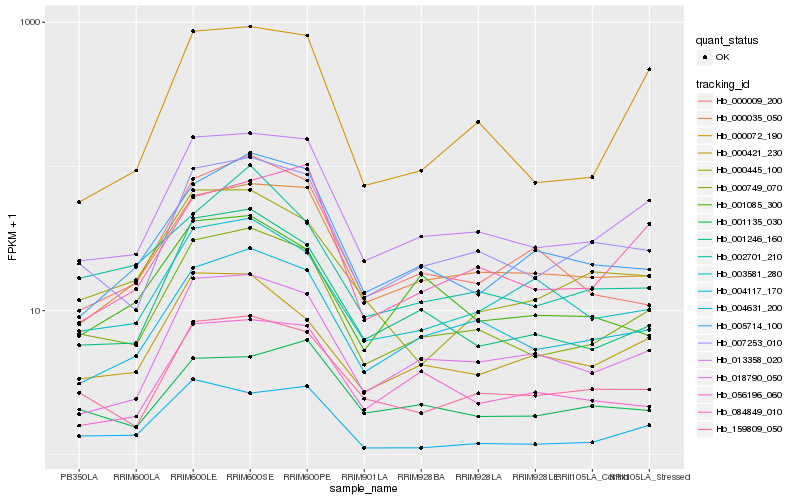
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_070 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 2 |
Hb_013358_020 |
0.0599865582 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 3 |
Hb_007253_010 |
0.0747069124 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_001246_160 |
0.0968824339 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 5 |
Hb_159809_050 |
0.0983684436 |
- |
- |
AT14A, putative [Ricinus communis] |
| 6 |
Hb_004117_170 |
0.0984009073 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 7 |
Hb_000035_050 |
0.1044049364 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 8 |
Hb_018790_050 |
0.1133690114 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 9 |
Hb_000421_230 |
0.1177742834 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 10 |
Hb_005714_100 |
0.1255936617 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 11 |
Hb_002701_210 |
0.1294016514 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_000072_190 |
0.1304563547 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 13 |
Hb_000445_100 |
0.1305274898 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 14 |
Hb_000009_200 |
0.1315099021 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 15 |
Hb_003581_280 |
0.1318396 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 16 |
Hb_004631_200 |
0.1365475863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_056196_060 |
0.1371954782 |
- |
- |
hexokinase [Manihot esculenta] |
| 18 |
Hb_084849_010 |
0.1372438513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001135_030 |
0.1401093479 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 20 |
Hb_001085_300 |
0.1409001389 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |