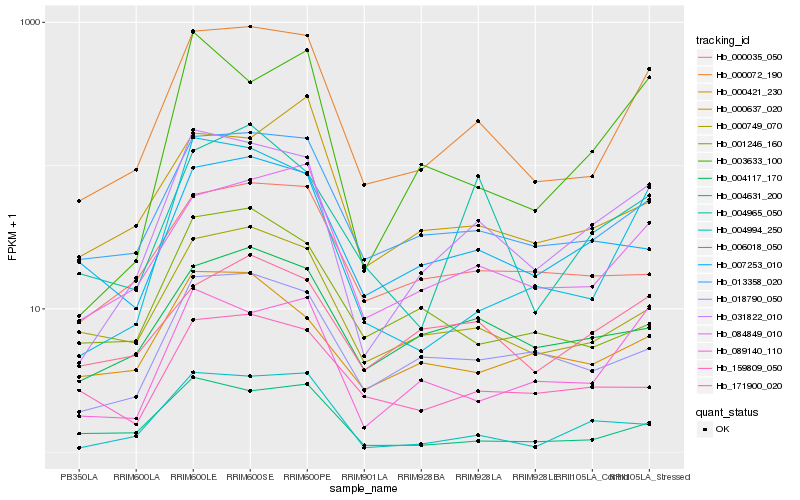
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_190 |
0.0 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 2 |
Hb_013358_020 |
0.1123405997 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 3 |
Hb_084849_010 |
0.1127932432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_031822_010 |
0.1224915755 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 5 |
Hb_000749_070 |
0.1304563547 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_006018_050 |
0.1406580039 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 7 |
Hb_004631_200 |
0.1487143577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_018790_050 |
0.1495673843 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 9 |
Hb_089140_110 |
0.1524061829 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004117_170 |
0.1601672319 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 11 |
Hb_007253_010 |
0.1663114691 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_004994_250 |
0.1684708965 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 13 |
Hb_000035_050 |
0.1705899496 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 14 |
Hb_171900_020 |
0.1752778057 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 15 |
Hb_004965_050 |
0.1769665109 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 16 |
Hb_000637_020 |
0.177350702 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 17 |
Hb_159809_050 |
0.1792024887 |
- |
- |
AT14A, putative [Ricinus communis] |
| 18 |
Hb_001246_160 |
0.1803311515 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 19 |
Hb_000421_230 |
0.1848573003 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 20 |
Hb_003633_100 |
0.1849098602 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |