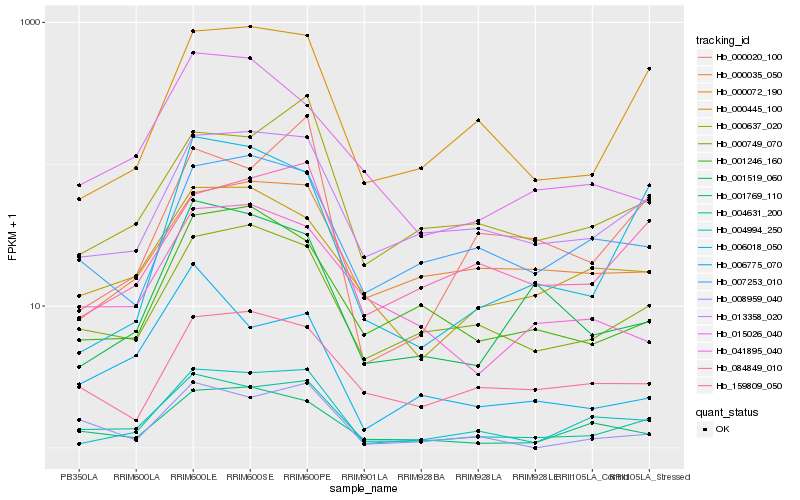
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000749_070 |
0.1365475863 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 3 |
Hb_013358_020 |
0.1371620148 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 4 |
Hb_000072_190 |
0.1487143577 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 5 |
Hb_004994_250 |
0.1530353768 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 6 |
Hb_000637_020 |
0.1551199229 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 7 |
Hb_000445_100 |
0.1578757776 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 8 |
Hb_008959_040 |
0.1633164584 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_001769_110 |
0.1639140755 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 10 |
Hb_084849_010 |
0.1651976196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001519_060 |
0.1677918391 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_007253_010 |
0.1730992702 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 13 |
Hb_001246_160 |
0.1753663299 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 14 |
Hb_159809_050 |
0.1761107265 |
- |
- |
AT14A, putative [Ricinus communis] |
| 15 |
Hb_041895_040 |
0.1780411404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000035_050 |
0.1806650471 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 17 |
Hb_006018_050 |
0.1811126512 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 18 |
Hb_006775_070 |
0.1819739997 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000020_100 |
0.1822937846 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 20 |
Hb_015026_040 |
0.1841349796 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |