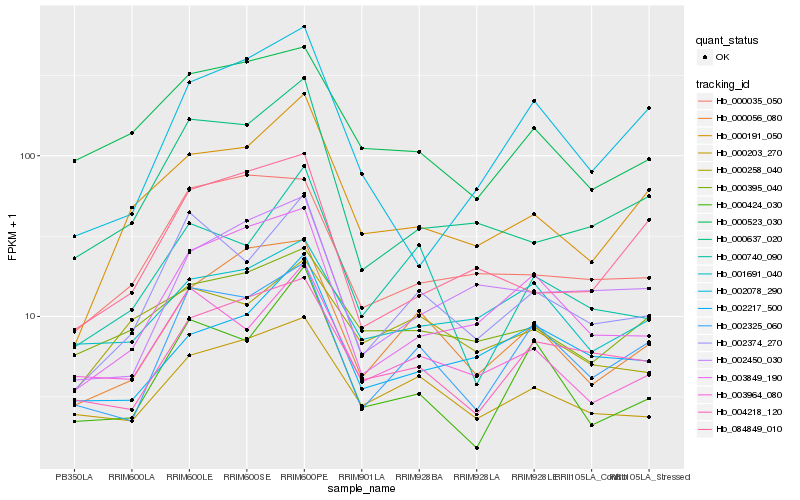
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 2 |
Hb_000637_020 |
0.1291871193 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 3 |
Hb_084849_010 |
0.1397240298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000395_040 |
0.1441267812 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 5 |
Hb_002374_270 |
0.1529584774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000523_030 |
0.1566539739 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 7 |
Hb_000056_080 |
0.1576078306 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003849_190 |
0.1600260811 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 9 |
Hb_003964_080 |
0.1649863728 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 10 |
Hb_000035_050 |
0.1678005751 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 11 |
Hb_002078_290 |
0.1681117249 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 12 |
Hb_002217_500 |
0.1710933088 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 13 |
Hb_002450_030 |
0.1714057044 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 14 |
Hb_000203_270 |
0.1734100119 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 15 |
Hb_000740_090 |
0.1743722518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004218_120 |
0.1747726612 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002325_060 |
0.1765672613 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 18 |
Hb_000258_040 |
0.1772654907 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 19 |
Hb_001691_040 |
0.1779664078 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000424_030 |
0.1816324615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |