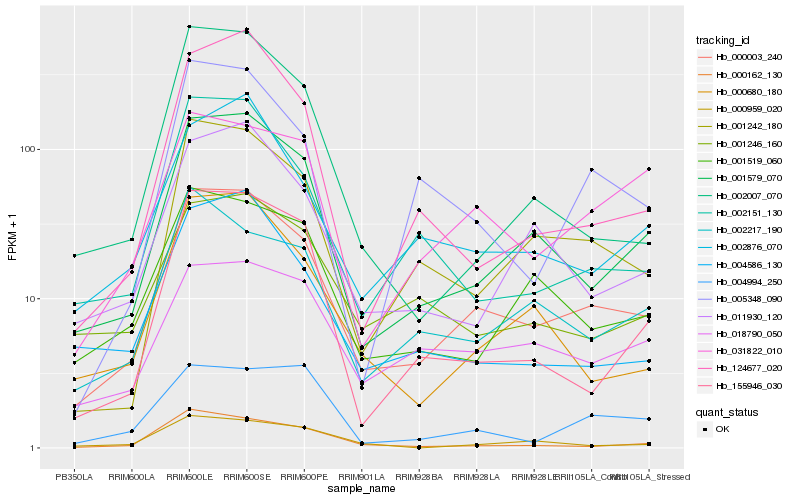
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001579_070 |
0.1095502851 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 3 |
Hb_001242_180 |
0.1295283861 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 4 |
Hb_000680_180 |
0.1458719698 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 5 |
Hb_002876_070 |
0.1472988332 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 6 |
Hb_001519_060 |
0.1475237435 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_011930_120 |
0.1479311667 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000162_130 |
0.1499110335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002151_130 |
0.1518035485 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 10 |
Hb_002217_190 |
0.1537605512 |
- |
- |
invertase [Hevea brasiliensis] |
| 11 |
Hb_004586_130 |
0.1577775357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_018790_050 |
0.1584124514 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 13 |
Hb_005348_090 |
0.1592884553 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 14 |
Hb_031822_010 |
0.1603492848 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 15 |
Hb_001246_160 |
0.1606317163 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 16 |
Hb_155946_030 |
0.1608086045 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 17 |
Hb_124677_020 |
0.1666891328 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 18 |
Hb_000959_020 |
0.1680975892 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 19 |
Hb_002007_070 |
0.1692042386 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 20 |
Hb_004994_250 |
0.1696868194 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |