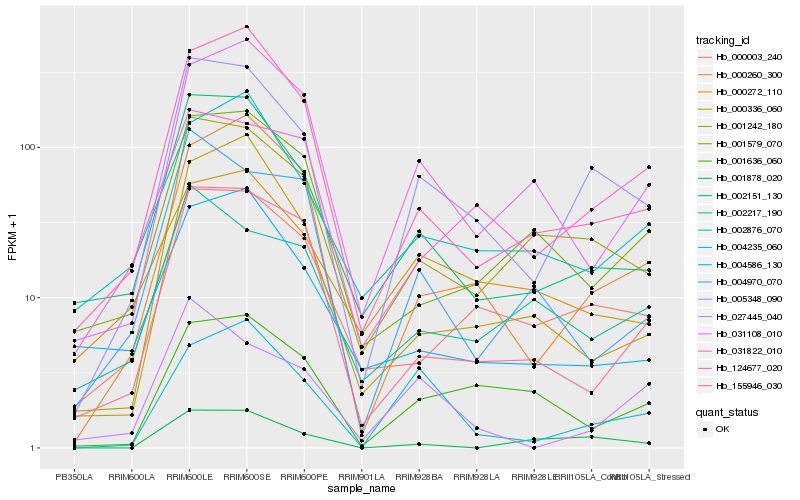
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005348_090 |
0.0 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 2 |
Hb_001242_180 |
0.1407970255 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 3 |
Hb_002151_130 |
0.145913424 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 4 |
Hb_000003_240 |
0.1592884553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000260_300 |
0.1720166797 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 6 |
Hb_124677_020 |
0.172451528 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_004235_060 |
0.1921910739 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_155946_030 |
0.1967657264 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 9 |
Hb_031108_010 |
0.1975300419 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 10 |
Hb_002876_070 |
0.1994151194 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 11 |
Hb_001878_020 |
0.1997273894 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 12 |
Hb_001579_070 |
0.2047982141 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 13 |
Hb_001636_060 |
0.2058969001 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 14 |
Hb_000336_060 |
0.2087470849 |
- |
- |
hypothetical protein JCGZ_26632 [Jatropha curcas] |
| 15 |
Hb_000272_110 |
0.2102991357 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002217_190 |
0.2126160101 |
- |
- |
invertase [Hevea brasiliensis] |
| 17 |
Hb_004586_130 |
0.2128504096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_027445_040 |
0.2140567006 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 19 |
Hb_004970_070 |
0.21696011 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 20 |
Hb_031822_010 |
0.2187870043 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |