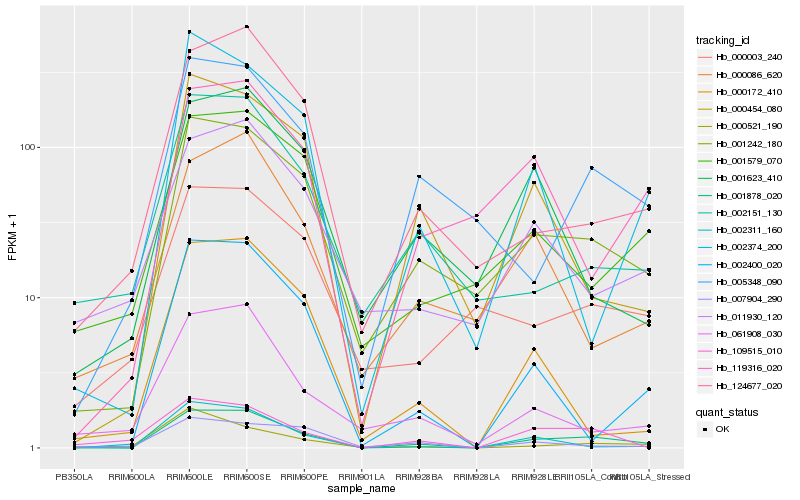
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001878_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 2 |
Hb_001242_180 |
0.1478809387 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 3 |
Hb_000454_080 |
0.1983963208 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 4 |
Hb_005348_090 |
0.1997273894 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 5 |
Hb_002400_020 |
0.2061949251 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 6 |
Hb_002311_160 |
0.209775495 |
transcription factor |
TF Family: ARR-B |
sensor histidine kinase, putative [Ricinus communis] |
| 7 |
Hb_124677_020 |
0.2100343045 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 8 |
Hb_002374_200 |
0.2116401511 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 9 |
Hb_001579_070 |
0.2200760437 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 10 |
Hb_119316_020 |
0.2223944686 |
- |
- |
- |
| 11 |
Hb_000172_410 |
0.2232132378 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_001623_410 |
0.2242500106 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_061908_030 |
0.2243134693 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 14 |
Hb_011930_120 |
0.2258464958 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007904_290 |
0.2277819183 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 16 |
Hb_000003_240 |
0.227892845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002151_130 |
0.2279382915 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 18 |
Hb_000521_190 |
0.228308195 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 19 |
Hb_109515_010 |
0.2284458647 |
- |
- |
- |
| 20 |
Hb_000086_620 |
0.2292034665 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |