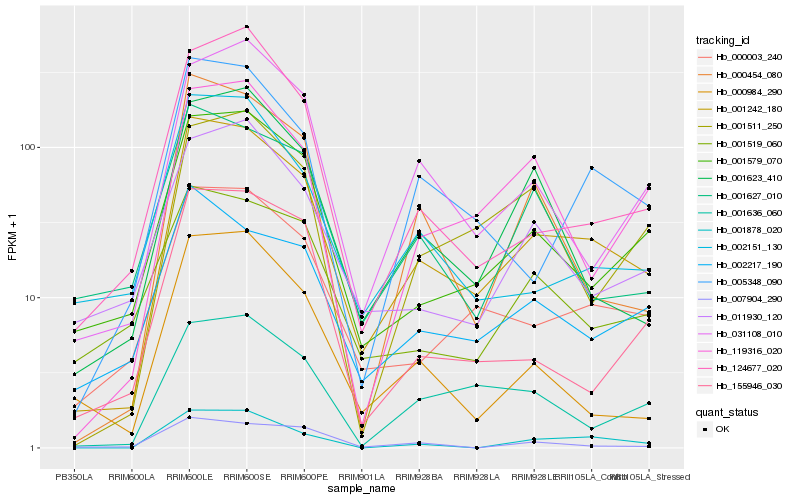
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001242_180 |
0.0 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 2 |
Hb_000003_240 |
0.1295283861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001579_070 |
0.1375407499 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 4 |
Hb_005348_090 |
0.1407970255 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 5 |
Hb_001878_020 |
0.1478809387 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 6 |
Hb_000454_080 |
0.1523786743 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 7 |
Hb_002217_190 |
0.1546567828 |
- |
- |
invertase [Hevea brasiliensis] |
| 8 |
Hb_031108_010 |
0.1564469729 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 9 |
Hb_001623_410 |
0.1579167018 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002151_130 |
0.158208944 |
- |
- |
12-oxophytodienoate reductase [Hevea brasiliensis] |
| 11 |
Hb_119316_020 |
0.1638085595 |
- |
- |
- |
| 12 |
Hb_001636_060 |
0.1651071315 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 13 |
Hb_001627_010 |
0.1658373715 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 14 |
Hb_011930_120 |
0.1661867766 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001519_060 |
0.1675351716 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_124677_020 |
0.1693261347 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 17 |
Hb_000984_290 |
0.1702661494 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_155946_030 |
0.1732948432 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 19 |
Hb_001511_250 |
0.1735822862 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 20 |
Hb_007904_290 |
0.1739181496 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |