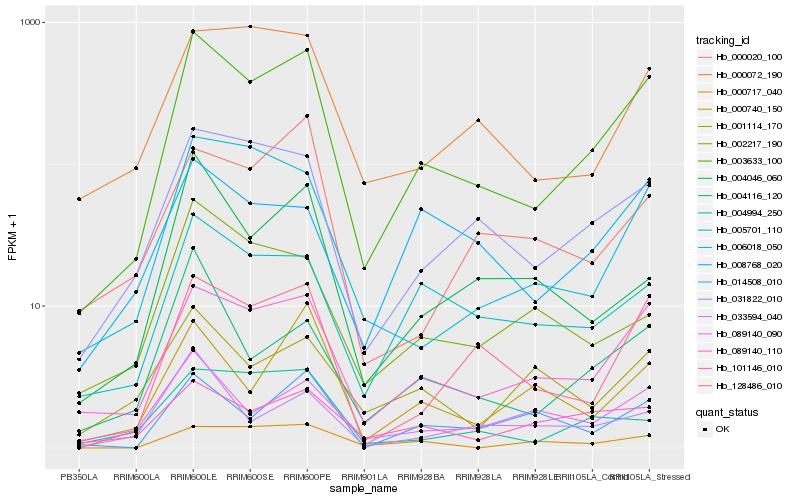
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003633_100 |
0.0 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 2 |
Hb_089140_110 |
0.1138905398 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_031822_010 |
0.1566781977 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 4 |
Hb_128486_010 |
0.1613538871 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 5 |
Hb_005701_110 |
0.1780829123 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 6 |
Hb_006018_050 |
0.1796583854 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 7 |
Hb_000072_190 |
0.1849098602 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 8 |
Hb_101146_010 |
0.1886387136 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 9 |
Hb_089140_090 |
0.191521141 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_004116_120 |
0.1952014727 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 11 |
Hb_004046_060 |
0.1997106323 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 12 |
Hb_004994_250 |
0.2029479941 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 13 |
Hb_014508_010 |
0.2088081294 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 14 |
Hb_000740_150 |
0.2089779009 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 15 |
Hb_000020_100 |
0.2107767141 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 16 |
Hb_033594_040 |
0.2126714204 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_008768_020 |
0.2136610241 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 18 |
Hb_002217_190 |
0.2151527267 |
- |
- |
invertase [Hevea brasiliensis] |
| 19 |
Hb_000717_040 |
0.216283766 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0015s03590g [Populus trichocarpa] |
| 20 |
Hb_001114_170 |
0.2177904245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |